Article • Rapid, reliable microbe identification
The Bologna Workflow System
Many countries across the world are challenged with a rising number of incidences of multi-drug resistant (MDR) organisms infecting the population, and for several years, a clear pattern of increased resistance has emerged in southern and eastern European countries.
Authors: Miriam Cordovana and Dr Markus Meyer
For example, in countries such as Italy, a reduced number of therapeutic options remain available for highly pathogenic infections, so research is targeting technologies that can rapidly detect infecting organisms with a high degree of accuracy, and establish the level of antimicrobial susceptibility.
A global health threat
Gram-negative bacteria are the most common cause of sepsis – a highly prevalent and deadly infection occurring worldwide – followed by Gram-positive bacteria and fungi. Fast isolation and species identification is therefore critical to provide a targeted therapeutic strategy and de-escalate from broad spectrum antibiotics as soon as possible. Sepsis is thought to impact approximately 27-30 million people globally every year [Source: Global Sepsis Alliance], with one third dying from the condition [Source: Vincent et al., 2014; Lancet Respiratory Medicine], so fast identification methods are required that allow clinicians to rapidly act upon results and manage blood stream infections.
Carbapenems are the most important family of last-line antibiotics for treating MDR Gram-negative bacterial infections, and carbapenem-resistant Enterobacteriaceae (CRE) have emerged relatively recently as a class of bacteria that are not susceptible to carbapenem treatment. Isolates that produce carbapenem-hydrolysing β-lactamases (carbapenemases) are the key target for studies to determine resistance mechanisms of CRE. Italy has one of the highest instances of carbapenem-resistant Klebsiella pneumonia in Europe, with 33.5% of invasive isolates showing resistance [Source: ECDC]. The Klebsiella pneumoniae Carbapenemase (KPC)-producing K. pneumoniae family is endemic in Italy and represents a severe public health concern, making rapid detection of these isolates a top priority.
Recommended article
Article • Infections
Carbapenem resistant strains
The increasing numbers of bacteria resistant to the newer generations of antibiotics is a public health problem on a global scale. Bacteria have an extraordinary capacity for adaptation, mutating permanently to overcome the action of our increasingly impotent antimicrobial armamentarium. A situation further aggravated by the use of the powerful ‘large spectrum’ antibiotics, creating further…
Until recently, techniques including the modified Hodge test, the disk-diffusion synergy test with inhibitors, and Carba NP (Nordmann Poirel) test, have either been slow (up to 24 hours) and time-consuming, lacking sensitivity, or have a limited number of targets included. Matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS) technology is now being recognised as a reliable, reproducible method to identify bacteria from positive blood cultures, and for the rapid detection of carbapenemase-producing bacterial strains.
Revolutionising the workflow

The hydrolytic activity of bacterial carbapenemases can now be detected by sensitive functional assays, based on the distinct mass changes of the carbapenem molecule after enzymatic cleavage. In contrast to previous methods, this MS-based phenotypic assay – Bruker’s MALDI Biotyper Selective Testing of Antibiotic Resistance-β-lactamase (MBT STAR-BL) – can be applied directly from positive blood cultures, after MBT Sepsityper isolation.
The S. Orsola-Malpighi University Hospital in Bologna, Italy, recently introduced the integrated Bologna Workflow to the Bacteriology, Mycology and Mycobacteriology section of the hospital’s Operative Unit of Microbiology, for rapid and cost-effective microbial identification and antimicrobial susceptibility testing (AST) from positive blood cultures. This is an urgent priority in clinical microbiology, as the spread of resistance against third-generation cephalosporins and carbapenems among Gram-negative bacteria poses a high burden on the Italian healthcare system, and is rapidly rising elsewhere in Europe and overall worldwide.
The Bologna Workflow consists of Bruker’s MALDI Biotyper platform, the MBT Subtyping Module, the MBT Sepsityper kit, and the MBT STAR-Carba and STAR-Cepha kits for fast phenotypic carbapenem- and cephalosporin-resistance testing.
Compared to traditional methods, which all require a subculturing step, the Bologna Workflow can save up to 24 hours, with a higher number of correct identifications and more samples identified. The previous subculture-based methods provided approximately 65% of correct identifications, compared to over 91% correct identifications with the Bologna Workflow.
Detecting resistant microbes
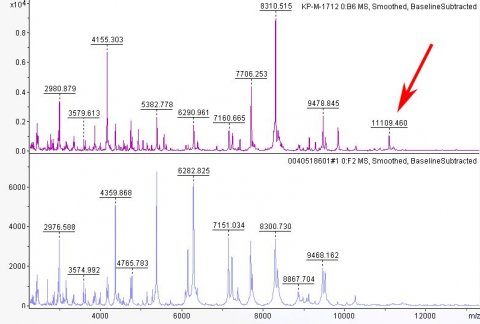
Reproduced from reference in accordance with the Creative Commons License.
KPC-producing K. pneumoniae first emerged in Italy in 2010 and since then, the Bologna hospital laboratory has been conducting a research project to phenotypically characterise and store the strains, and evaluate different methods of detection. Lau et al. (2014) discovered a specific MALDI-TOF mass peak at 11,109 m/z appearing in KPC-producing isolates, which is related to the pKpQIL plasmid carrying the blaKPC gene [Source: Lau et al., 2014; Journal of Clinical Microbiology].
The automated identification of these KPC-producers, by detecting this specific KPC-related peak in the MALDI-TOF MS spectra, is now implemented in the MBT Subtyping Module, allowing a “presumptive KPC” result (Figure 1 and Figure 2). This innovative approach was applied first to bacterial plate cultures, then directly to positive blood culture bottles, using the bacterial pellet obtained using the MBT Sepsityper, which enabled reliable and rapid identification of KPC-producing K. pneumoniae strains. The automated peak detection allowed for 100% specificity and 85.1% sensitivity. In addition to the subtyping, the STAR-Carba test for confirmation of carbapenemase production showed 100% specificity and sensitivity [Source: Cordovana et al, 2018; Frontiers in Microbiology].
The Bologna group is achieving a turnaround time for detection of KPC-producing isolates of 10 min to 1.5 hours, and showed that this novel MALDI-based approach uniquely provides real time detection of an antibiotic resistance marker, simultaneously with species identification.
Resistance detection in the future
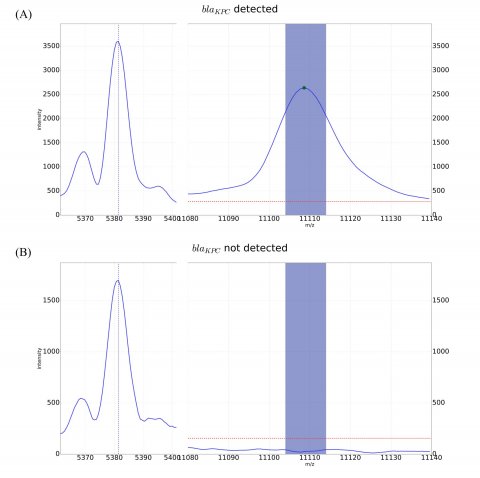
Reproduced from reference in accordance with the Creative Commons License.
By incorporating the MALDI MS-based approach the laboratory at the S. Orsola-Malpighi University Hospital can perceive early warnings for KPC-producing strains, facilitating the rapid initiation or change of therapeutic action and future infection control measures.
The future will see a wide scope for the extension of KPC-subtyping, as well as for the addition of new resistance markers and resistance detection methods. There is an increasing need for more 'customised' susceptibility testing, tailored by the results of species identification, combined with the epidemiological data of each setting. The ever-increasing spread of uncommon opportunistic pathogens and the growing proportion of immunocompromised patients for specific pathologies or medical treatments, is of significant concern. In the future this could present a further challenge to AST. Consequently, the faster microbiologists can identify the organisms, the stronger the overall patient outcomes will be.
Profiles:
Miriam Cordovana is a Biomedical Laboratory Technician at the S. Orsola-Malpighi University Hospital, which she joined in May 2007, following roles at the University Hospital of Florence Careggi and the Human Genetic Laboratory of the University of Florence. Miriam oversees the processing of biological samples, from their arrival at the laboratory, through identification to the execution of antimicrobial susceptibility testing. She has played an integral role in the department’s aim to optimise the entire workflow for blood culture microbial testing.
Dr Markus Meyer is a Global Product Manager at Bruker Daltonics. He gained his PhD in cell biology, biochemistry and microbiology at the University of Münster, Germany, and joined Bruker in 2005 as an Application Specialist in Clinical Proteomics.
S. Orsola-Malpighi University Hospital
More than 400 years ago the Bologna University Hospital Authority St. Orsola-Malpighi Polyclinic became the first hospital in Bologna. Today it is home to the School of Medicine and Surgery. The Polyclinic, an internationally acclaimed institution for the study and treatment of pathologies, organises annual medical conferences and conventions attended by professionals of international fame. The hospital develops and delivers multi-specialist research, education and training in an integrative manner, and promotes innovation and pursues the highest degree of patient/customer care and the training of medical students, as well as expanding the full potential of its skilled staff through goal sharing and assuming responsibility for any results.
The Bruker Corporation
‘Bruker’s high-performance scientific instruments and high-value analytical and diagnostic solutions enable scientists to explore life and materials at molecular, cellular and microscopic levels. In close cooperation with our customers, Bruker is enabling innovation, improved productivity and customer success in life science molecular research, in applied and pharma applications, in microscopy and nanoanalysis, and in industrial applications, as well as cell biology, preclinical imaging, clinical phenomics and proteomics research and clinical microbiology.’
For more details, please visit: www.bruker.com.
More information on Bruker’s MALDI Biotyper systems can be found at: https://www.bruker.com/products/mass-spectrometry-and-separations/maldi-biotyper-systems
03.07.2019