Image source: Adobe Stock/mi_viri
News • Oncology & health informatics
Colon cancer: AI tool predicts survival, treatment response
A new artificial intelligence model designed by researchers at Harvard Medical School and National Cheng Kung University in Taiwan could bring much-needed clarity to doctors delivering prognoses and deciding on treatments for patients with colorectal cancer, the second deadliest cancer worldwide.
Solely by looking at images of tumor samples—microscopic depictions of cancer cells—the new tool accurately predicts how aggressive a colorectal tumor is, how likely the patient is to survive with and without disease recurrence, and what the optimal therapy might be for them.
A report on the team's work is published in Nature Communications.
Having a tool that answers such questions could help clinicians and patients navigate this wily disease, which often behaves differently even among people with similar disease profiles who receive the same treatment—and could ultimately spare some of the 1 million lives that colorectal cancer claims every year.
The researchers caution that the tool is meant to enhance, not replace, human expertise.
"Our model performs tasks that human pathologists cannot do based on image viewing alone," said study co-senior author Kun-Hsing Yu, assistant professor of biomedical informatics in the Blavatnik Institute at HMS. Yu led an international team of pathologists, oncologists, biomedical informaticians, and computer scientists.
"What we anticipate is not a replacement of human pathology expertise, but augmentation of what human pathologists can do," Yu added. "We fully expect that this approach will augment the current clinical practice of cancer management."
Patient's prognosis depends on multiple factors
The researchers caution that any individual patient's prognosis depends on multiple factors and that no model can perfectly predict any given patient's survival. However, they add, the new model could be useful in guiding clinicians to follow up more closely, consider more aggressive treatments, or recommend clinical trials testing experimental therapies if their patients have worse predicted prognoses based on the tool's assessment.
The tool could be particularly useful in resource-limited areas both in this country and around the world where advanced pathology and tumor genetic sequencing may not be readily available, the researchers noted.
The new tool goes beyond many current AI tools, which primarily perform tasks that replicate or optimize human expertise. The new tool, by comparison, detects and interprets visual patterns on microscopy images that are indiscernible to the human eye.
The tool, called MOMA (for Multi-omics Multi-cohort Assessment) is freely available to researchers and clinicians.
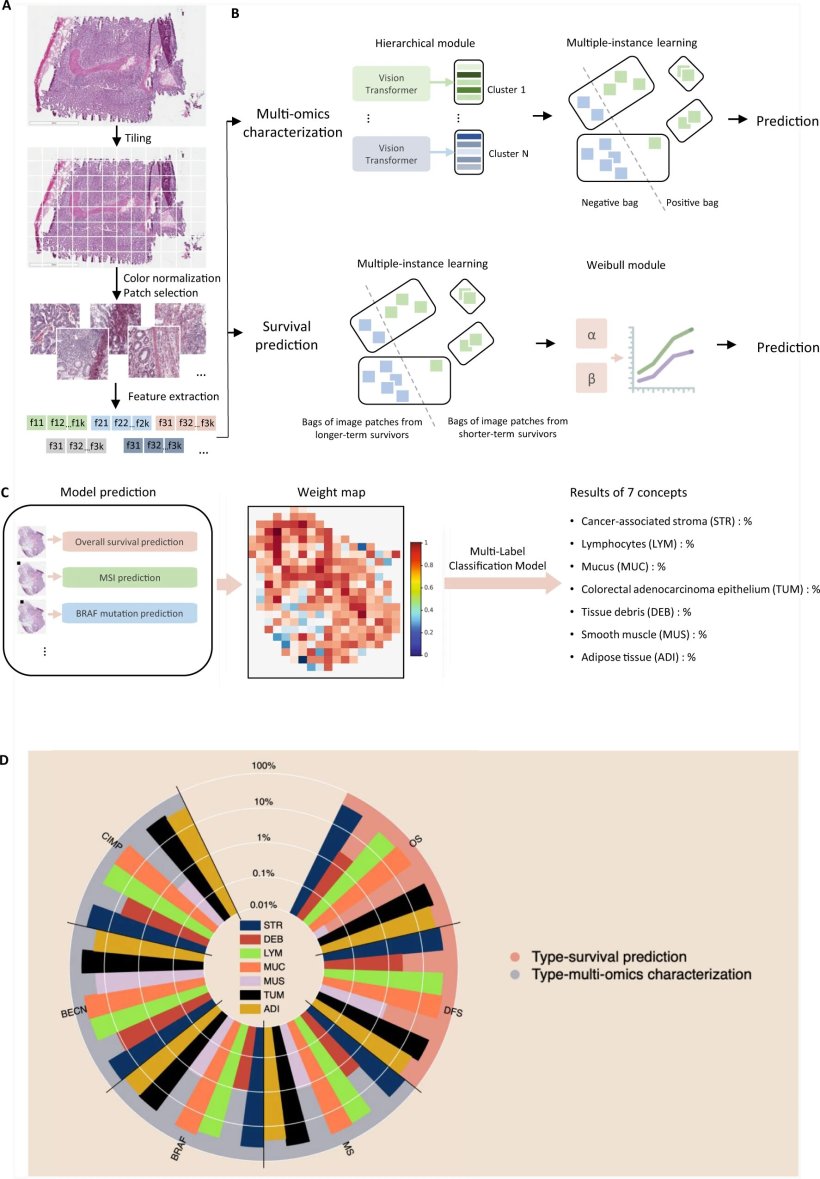
Image source: Nature Communications (2023). DOI: 10.1038/s41467-023-37179-4
Detailed description of the MOMA machine learning framework
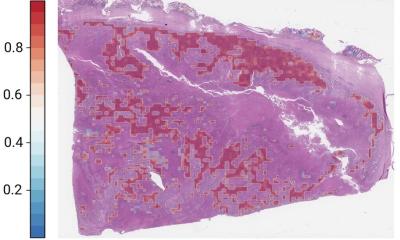
A Machine learning workflow for connecting whole-slide digital histopathology images with multi-omics biomarkers and survival outcomes. The MOMA platform processes the image patches from whole-slide pathology images, normalizes them, and leverages vision transformers to extract image features.
B We develop multi-omics characterization and survival prediction frameworks using the extracted image features.
C Model visualization and interpretation. To enhance the interpretability of our machine learning approaches, we compute the importance of each image region to the prediction target by quantifying the performance decay due to occlusion of the region, and we develop a multi-task classification model to quantify the concept (e.g., lymphocyte, stroma, tumor, adipose tissue, mucin, etc.) score using patches whose importance weight is greater than 0.7. This method connects prior histopathology knowledge with quantitative importance metrics independently learned by the models.
D A summary of the pathology concepts associated with survival and multi-omics predictions. The concept scores are plotted on the log scale. OS: overall survival prediction in early-stage CRC; DFS: disease-free survival prediction in early-stage CRC; MSI: microsatellite instability prediction; BRAF: BRAF mutation status prediction; BECN: BECN-1 overexpression prediction; CIMP: CpG island methylator phenotype prediction. The major concepts visualized here include lymphocytes (LYM), cancer-associated stroma (STR), tissue debris (DEB), mucus (MUC), smooth muscle (MUS), colorectal adenocarcinoma epithelium (TUM), and adipose tissue (ADI). The score for each concept indicates the relative importance of each type of microenvironment in predicting patient prognoses or the selected multi-omics variations with clinical implications.
Extensive training and testing
It's important to augment the model with new and more data as they come along so that its performance never lags behind.
Kun-Hsing Yu
The model was trained on information obtained from nearly 2,000 patients with colorectal cancer from diverse national patient cohorts that together include more than 450,000 participants—the Health Professionals Follow-up Study, the Nurses' Health Study, the Cancer Genome Atlas Program, and the NIH's PLCO (Prostate, Lung, Colorectal and Ovarian) Cancer Screening Trial.
During the training phase, the researchers fed the model information about the patients' age, sex, cancer stage, and outcomes. They also gave it information about the tumors' genomic, epigenetic, protein, and metabolic profiles.
Then the researchers showed the model pathology images of tumor samples and asked it to look for visual markers related to tumor types, genetic mutations, epigenetic alterations, disease progression, and patient survival.
The researchers then tested how the model might perform in "the real world" by feeding it a set of images it had not seen before of tumor samples from different patients. They compared its performance with the actual patient outcomes and other available clinical information.
The model accurately predicted the patients' overall survival following diagnosis, as well as how many of those years would be cancer-free. The tool also accurately predicted how an individual patient might respond to different therapies, based on whether the patient's tumor harbored specific genetic mutations that rendered the cancer more or less prone to progression or spread. In both of those areas the tool outperformed human pathologists as well as current AI models. The researchers said the model will undergo periodic upgrading as science evolves and new data emerge.
"It is critical that with any AI model, we continuously monitor its behavior and performance because we may see shifts in the distributions of disease burden or new environmental toxins that contribute to cancer development," Yu said. "It's important to augment the model with new and more data as they come along so that its performance never lags behind."
Recommended article

News • Reduced postoperative complications
Colorectal cancer surgery: gut microbiota helps healing
In a promising study, Canadian researchers have shown for the first time in mice that modifying intestinal flora before surgery could reduce postoperative complications in colorectal cancer patients.
Discerning telltale patterns
The new model takes advantage of recent advances in tumor imaging techniques that offer unprecedented levels of detail, which nonetheless remain indiscernible to human evaluators. Based on these details, the model successfully identified indicators of how aggressive a tumor was and how likely it was to behave in response to a particular treatment.
Based on an image alone, the model also pinpointed characteristics associated with the presence or absence of specific genetic mutations—something that typically requires genomic sequencing of the tumor. Sequencing can be time-consuming and costly, particularly for hospitals where such services are not routinely available.
It is precisely in such situations that the model could provide timely decision support for treatment choice in resource-limited settings or in situations where there is no tumor tissue available for genetic sequencing, the researchers said.
The researchers said that before deploying the model for use in clinics and hospitals, it should be tested in a prospective, randomized trial that assesses the tool's performance in actual patients over time after initial diagnosis. Such a study would provide the gold-standard demonstration of the model's capabilities, Yu said, by directly comparing the tool's real-life performance using images alone with that of human clinicians who use knowledge and test results that the model does not have access to.
Another strength of the model, the researchers said, is its transparent reasoning. If a clinician using the model asks why it made a given prediction, the tool would be able to explain its reasoning and the variables it used.
This feature is important for increasing clinicians' confidence in the AI models they use, Yu said.
Gauging disease progression, optimal treatment
The model accurately pinpointed image characteristics related to differences in survival. For example, it identified three image features that portended worse outcomes:
- Greater cell density within a tumor.
- The presence of connective supportive tissue around tumor cells, known as stroma.
- Interactions of tumor cells with smooth muscle cells.
The model also identified patterns within the tumor stroma that indicated which patients were more likely to live longer without cancer recurrence. The tool also accurately predicted which patients would benefit from a class of cancer treatments known as immune checkpoint inhibitors. While these therapies work in many patients with colon cancer, some experience no measurable benefit and have serious side effects. The model could thus help clinicians tailor treatment and spare patients who wouldn't benefit, Yu said.
The model also successfully detected epigenetic changes associated with colorectal cancer. These changes—which occur when molecules known as methyl groups attach to DNA and alter how that DNA behaves—are known to silence genes that suppress tumors, causing the cancers to grow rapidly. The model's ability to identify these changes marks another way it can inform treatment choice and prognosis.
Source: Harvard Medical School
18.04.2023