News • Patient-derived whole-tumor cell culture model
Breast cancer: New method finds the right therapy
Researchers at Karolinska Institutet have developed a method that should be able to predict whether a patient with breast cancer will benefit from a particular treatment or not.

Image source: Karolinska Institutet; photo: Stefan Zimmerman
The cell-based method has been tested on patients with promising results, according to a study published in PNAS.
“Today, there are limited possibilities of determining in advance which breast cancer patients benefit from different treatments. This method can predict how patients will respond to certain treatments, which means that unnecessary side effects can be avoided, and costs can be saved. Larger confirmatory studies are needed, but we see that the concept works,” says Johan Hartman, professor at the Department of Oncology-Pathology, Karolinska Institutet, and the study’s corresponding author.
Several different drugs are currently available for the treatment of breast cancer. But the disease is complex and not all patients are helped by the same or all drugs. Some information about which treatment a patient will benefit from can be obtained through, for example, DNA sequencing. However, in many cases, it is not possible to say whether a treatment will help the individual patient or not, and therefore better methods are needed to predict how patients will respond to treatment.
The method the KI researchers have developed is based on isolating and cultivating not only tumour cells but also so-called supporting cells from patients with breast cancer. The cancer treatments are then tested in different concentrations on the cultured cells to see how sensitive they are to the drugs.
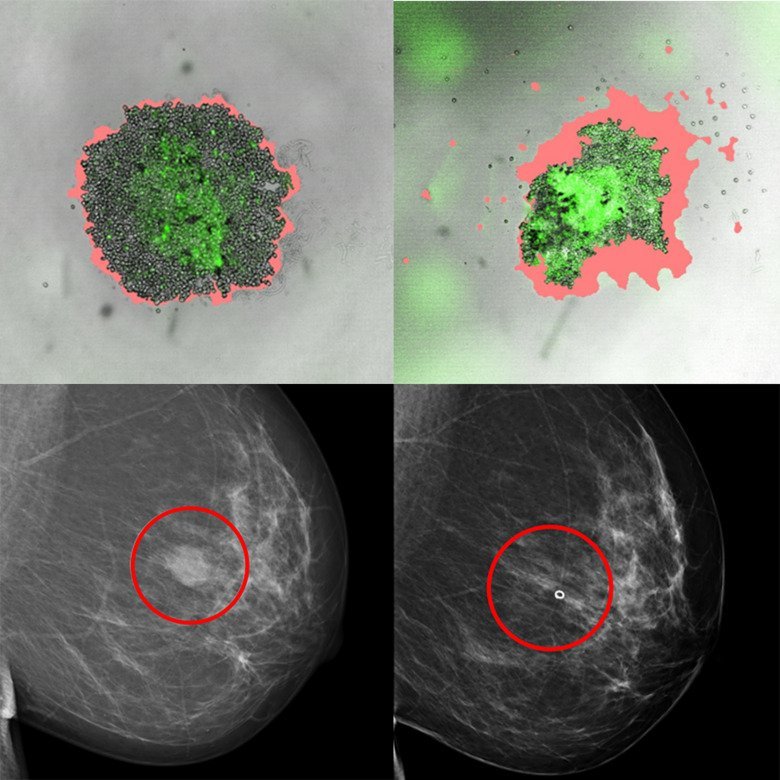
Image source: Chen et al., PNAS 2022 (CC BY 4.0)
In the current study, the researchers show that it is possible to establish this type of cell-based tumour model from breast tumours and that the cell models are similar to the patients’ tumours of origin in relevant ways, for example genetically and in terms of different protein markers. The tumour models were created from biopsies from 98 patients who had undergone breast cancer surgery. More than 35 existing breast cancer drugs and breast cancer drugs under development were tested on them. The researchers could then see that the drug sensitivity the tumour models displayed was very much consistent with current knowledge regarding treatment options based on patient tumour types and that in some cases the tumour models were sensitive to the drugs that were under development.
Next, the researchers examined how accurately the method can predict treatment responses. A validation study was carried out on 15 breast cancer patients who received treatment before surgery, so-called neoadjuvant treatment, at Södersjukhuset in Stockholm. From biopsies taken before surgery, the researchers created tumour models for each patient and exposed them to the same drugs the patient received. Thereafter, the drug sensitivity of the models was compared with the patients’ treatment response.
Image source: Karolinska Institutet; photo: Niklas Elmehed
The results show that the treatment responses predicted by the tumour model were broadly consistent with the treatment responses the patient subsequently exhibited. For example, the model predicted the treatment response from the chemotherapy drug epirubicin with 90% accuracy, while four of four patients treated and tested for anti-HER2 monoclonal antibody drugs showed consistency.
The researchers note it is also possible to create cell models from small tumours, which are often considered technically challenging to sample without jeopardizing diagnostics, and that the tests can be carried out relatively quickly. “In most cases, we can perform individualized drug testing and get the result within ten days, indicating that this method can work in daily clinical practice. But it can also be used in research and drug development,” says the study's first author Xinsong Chen, research specialist at the Department of Oncology-Pathology, Karolinska Institutet.
The next step is to test the method in a larger group of patients and to investigate the possibility of combining it with other molecular methods for an even better prediction of treatment responses as well as study resistance mechanisms.
The research is funded by the Swedish Cancer Society, the Swedish Breast Cancer Association, Region Stockholm, Radiumhemmet's research funds, and the Swedish Research Council. Johan Hartman is co-founder and co-owner of Stratipath, which develops methods for AI-based decision support for cancer diagnostics. However, the company has no connection to the current study. Outside of this study, some of the researchers have received reimbursements from pharmaceutical companies. See the scientific study for more information.
Source: Karolinska Institutet
03.01.2023