Image source: Kim T et al., Journal of Medical Imaging 2024 (CC BY 4.0)
News • Color adjustment technique
More consistent histopathology slides for machine learning use
In histopathology, where tissues are studied under the microscope to understand and diagnose diseases, stains represent a quintessential tool.
Simply put, stains are carefully selected or crafted chemicals that adhere to specific cellular components. When viewed under a microscope, they help the user distinguish cell structures more easily by altering the observed colors.
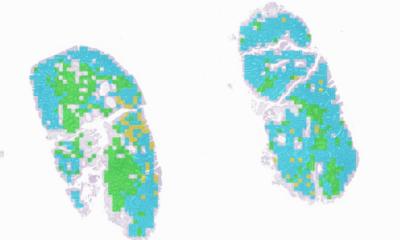
Datasets of stained images depicting both normal and diseased tissues are valuable for training machine learning models, which can help doctors assess tricky cases and mitigate personal biases during diagnosis. To ensure these models work properly, it is important to minimize color differences between the images used for training and those they will analyze in real-world scenarios. Using so-called “domain adaptation techniques,” variations in color that originate from the unique experimental setups used in different laboratories can be corrected, creating more consistent and comparable data.
In a recent study published in Journal of Medical Imaging (JMI), researchers from University of North Carolina at Chapel Hill (United States) proposed a novel domain adaptation technique. Termed Stain simultaneous augmentation and normalization (Stain SAN), the proposed method can help make stained histopathological image datasets more useful for many emerging machine-learning-based classification systems, ultimately leading to improved diagnostic tools.
Stain SAN achieves results comparable with a state-of-the-art deep learning-based approach without requiring separate training for stain adaptation or access to the target domain during training, which would be unrealistic in clinical practice
Taebin Kim
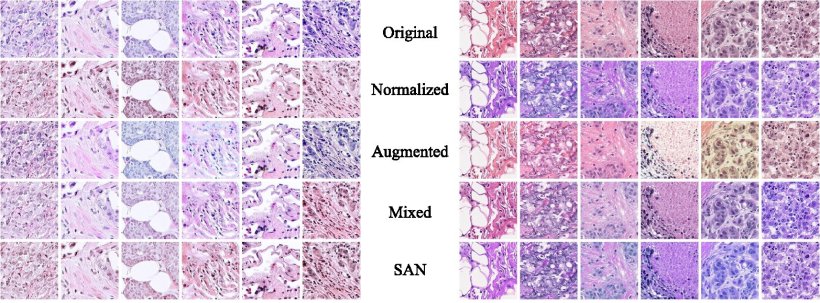
Stain SAN involves three main steps:
- stain extraction,
- color adaptation, and
- intensity adaptation.
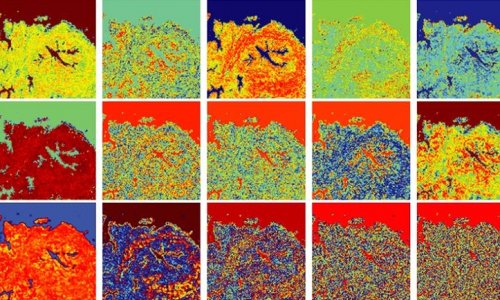
In the first step, the original stain image is decomposed into a product of two matrices: one containing color information and the other containing light intensity information for each pixel. In the adaptation step, the distribution of colors in the color matrix is modified through a statistical process that considers the training images, guaranteeing that the altered colors fall in a target distribution. Finally, in the third step and before image reconstruction, the intensity matrix undergoes random perturbation. This helps increase the diversity of possible stain domains.
“The main benefit of Stain SAN is that it combines the strengths of previous stain adaptation methods while overcoming their inherent weaknesses,” explains Dr. Taebin Kim, the lead researcher. He further adds, “Other established techniques, including stain normalization, stain augmentation, and stain mix-up, can be understood as special cases of Stain SAN.”
The researchers tested their approach, both qualitatively and quantitatively, using histopathological images from publicly available datasets. Based on their observations as well as feedback from an expert pathologist, the researchers found that image datasets processed using Stain SAN led to more consistently aligned colors, with better generalized stain domains. Moreover, Stain SAN increased the contrast between the nucleus and cytoplasm in each cell and highlighted differences between tumor cells and supportive tissue.
The team also trained machine-learning-based classifiers using datasets processed via various domain adaptation techniques and tested their performance against processed images from another dataset. Interestingly, Stain SAN outperformed all the aforementioned methods, delivering markedly higher accuracy. “Our results clearly show the improvements made over the history of the development of these methods, culminating with a substantial enhancement provided by Stain SAN,” comments Kim. But that is not all, as he adds, “Furthermore, we showed that Stain SAN achieves results comparable with a state-of-the-art deep learning-based approach without requiring separate training for stain adaptation or access to the target domain during training, which would be unrealistic in clinical practice. This highlights the effectiveness and computational efficiency of Stain SAN.”
The development of efficient domain adaptation techniques like Stain SAN is essential to bridge the gap that exists between machine learning systems and their applications in healthcare. The research team is already planning on potential improvements to their method and conducting further testing using other datasets. “Our findings endorse Stain SAN as a robust approach for stain domain adaptation in histopathology images, with implications for advancing computational tasks in the field,” concludes Kim, optimistic about the future. Their efforts will pave the way to more accurate and convenient diagnostic protocols, saving time for doctors and patients alike.
Source: SPIE--International Society for Optics and Photonics
30.08.2024