Sex-linked disorders
X-Citing X Chromosome
A team of scientists from the University of Michigan Medical School shows that the genetic material in female (but not male) cells makes tiny amounts of a special genetic material called RNA to make one of the two X chromosomes silent. They call this RNA XistAR.
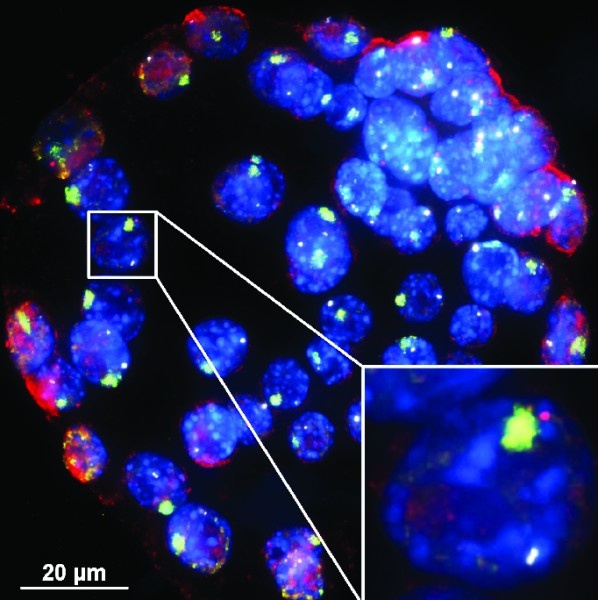
Think back to middle school biology class, when you learned that boys have an X and a Y chromosome inside each cell, and girls have two X’s. That small difference explains why boys are more at risk for a range of disorders -- from autism to hemophilia -- linked to genetic defects on their lone X chromosome. Meanwhile, most girls who carry the same defect on one X chromosome don’t get sick, because their other X chromosome runs the show.
Now, a new genetic discovery could help scientists understand exactly how this silencing of one X happens – and perhaps lead to better treatment for X-linked diseases. Back in biology class, you probably learned that RNAs make proteins in cells. But XistAR is special. It belongs to a class of RNAs called lncRNAs, or long noncoding RNAs, that don’t make proteins, but instead control how our genetic code gets read.
X marks the spot
The question that U-M geneticist Sundeep Kalantry, Ph.D., and his team set out to answer was: how do female cells turn off one of their two X chromosomes? They knew that a gene called Xist causes cells to make a kind of lncRNA that coats the “silenced” X and keeps it dormant. But how is Xist lncRNA itself made?
Genes can be ‘read’ in both the forward and the backward direction. The Kalantry team found that the Xist gene on the otherwise “silent” X chromosome is read in both the forward and, unexpectedly, in the backward direction. This makes the RNA they dubbed XistAR, short for ‘Xist Activating RNA’. Production of XistAR in the backward direction is required to generate Xist RNA in the forward direction and turn off the X chromosome.
To prove the discovery, the team had to develop a new kind of test to detect such RNA strands, which cells make in much smaller amounts than the forward-reading variety. The test could now help scientists look at other places in the genome for more examples of rare lncRNAs -- made in the backward or ‘antisense’ direction -- that regulate the genetic code.
The work was done in mice, which allowed the scientists to see what happened when they kept cells from making XistAR. Without it, the cells couldn’t silence one X chromosome effectively, and they made too much of the X-chromosome gene products in females.
Promise for the future
If the discovery translates to humans – and XistAR seems to also be present in humans -- Kalantry believes that XistAR may provide a convenient handle to manipulate X chromosome genes. For example, it could be possible to boost proteins generated by the X chromosome’s genes by stopping XistAR from working. He and his team are now looking for factors that control XistAR activity.
“This work sheds light into how lncRNAs function, how genes and even an entire chromosome can be quieted. XistAR provides a molecular target to control gene expression – how to ‘wake the genes up’ or reduce their activity,” says Kalantry. “Exploring how the X chromosome becomes inactivated lets us know how to selectively activate it. Turning on the healthy copy of an X chromosome gene maybe a way to minimize disease risks associated with the X chromosome.” Meanwhile, he says, understanding how antisense lncRNAs can work in an “epigenetic” way – acting as an outside force on DNA to control whether it gets read or not – can apply far beyond the X-chromosome.
Calico cats and autism
Kalantry, an assistant professor in the U-M Department of Human Genetics, keeps a portrait of a calico cat on his office wall. It reminds him of the power of X-chromosome inactivation. In such cats, nearly all of them female, patches of fur of different colors grow from different regions of skin. The color of each patch depends on which X-chromosome has been turned off and which is on in those hair-producing cells – whether it’s the one inherited from the cat’s mother or the one from the father.
X-inactivation, also called Lyonization, happens in the earliest stages of a female embryo’s development. Once each cell in the tiny clump of embryonic cells has “decided” which X to silence, all the cells that ever develop from that cell also inactivate copies of the same X.
Unlike calico cats, however, the effects of inactivating the “healthy” X chromosome more often than the “unhealthy” one can really change a girl’s life. For example, autism is heavily linked to genetic problems on the X-chromosome, which explains why boys, with only one X chromosome, are more than four times as likely as girls to fall on the autistic spectrum. Another autism-like disorder, called Rett syndrome, occurs only in girls, when a mutated gene on one of their X chromosomes gets used.
Next steps
While the new discovery does not immediately show a path to treating autism or Rett syndrome by altering activity of “unhealthy” X chromosomes, it does pave the way for further research. Strategies to block RNA strands in cells are already being tested in clinical trials.
Kalantry notes that the new work immediately opens up ways to find short-lived lncRNAs such as XistAR. The team developed ways to “tag” such strands to make them detectable.
“The control of genes by lncRNAs, often via epigenetic means, is now appreciated to occur in a wide variety of contexts, from normal physiology to diseases. On a fundamental level, it controverts the central dogma of DNA begetting RNA, which then makes proteins,” says Kalantry. “The techniques we’ve developed facilitate the discovery of rare RNA species in a cell. Such RNAs have been missed by high-throughput sequencing approaches, but maybe essential for cell function.”
Source: University of Michigan Health System
06.11.2015