Article • dPCR and HSAFM
Low-cost technique for missed genetic mutations
A new low-cost method targeting genetic mutations often missed by existing diagnostic approaches has been developed. Researchers at Virginia Commonwealth University (VCU) in the United States noted that most rearrangement mutations implicated in cancer and neurological diseases fall between what can be detected by DNA sequence reads and optical microscopy methods.
Report: Mark Nicholls
Image source: PublicDomainPictures from Pixabay
The new technique combines digital polymerase chain reaction (dPCR) and high-speed atomic force microscopy (HSAFM) to create an image at a nanoscale resolution, which allows technicians to measure differences in the lengths of genes and identify these missing mutations.

Dr Jason Reed, an associate professor in the department of physics at the VCU College of Humanities and Sciences, said: “Our approach is the first to combine dPCR with HSAFM to create an image with such nanoscale resolution that users can measure differences in the lengths of genes in a DNA sequence. These variations in gene length, known as polymorphisms, can be key to accurately diagnosing many forms of cancer and neurological diseases.”
The dPCR uses the DNA polymerase enzyme to exponentially clone samples of DNA or RNA for further experimentation or analysis. The sample is then placed on an atomically flat plate for inspection using HSAFM, which drags a sharp microscopic stylus across the sample to create precise measurements at a molecular level. “Our technique,” he continued, “uses easily accessible and affordable optical lasers, like those in a DVD player, to process samples at a rate thousands of times faster than typical atomic force microscopy.” From there, the team developed computer codes to trace the length of each DNA molecule.
Dr Reed said genetic mutations can be missed by existing diagnostic approaches as DNA-length polymorphisms are difficult to diagnose due to their repetitive or variable elements. For example, within the FLT3 gene, internal tandem duplications (ITDs) range from <30 bp to >200 bp (base pairs) in length and are associated with poor prognosis in acute myeloid leukaemia.
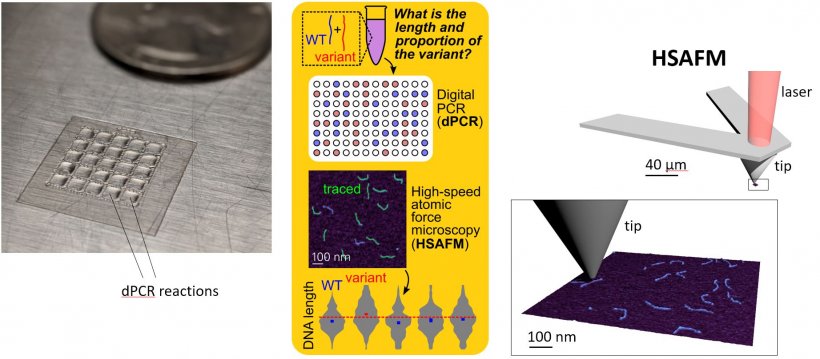
Image cortesy of Dr Jason Reed
The VCU team believe their approach could have applications in a number of areas of medicine. Dr Reed explained that many forms of cancer and inherited neurological diseases involve insertions or deletions of DNA material that can be difficult to characterize with DNA sequencing. The current standard test for diagnosing FLT3 gene mutations is the Leukostrat CDx FLT3 mutation assay. “Our approach shows variant allele fraction, which the current approach does not, and we are able to stitch together multiple images to show the lengths of extremely long molecules,” he added. “The speed, low cost, and flexibility of dPCR–HSAFM to detect long, repetitive insertions (>200 bp) make it an attractive alternative to sequencing or other more costly assays in the clinical diagnosis of length polymorphisms.”
Clinicians and researchers who work on neurological disorders like Huntington’s disease, Fragile X syndrome, movement disorders such as Parkinson’s, have already expressed interest.
If commercialized, this process could offer a significantly cheaper alternative for detecting and diagnosing DNA length polymorphisms
Jason Reed
For patients, the new approach could lead to more accurate diagnoses and the potential to diagnose future diseases distinguished by DNA length polymorphisms. But a big appeal is that it is fast and cost-effective and at less than $1 per sample to scan as dPCR is a common and affordable process for amplifying genetic material for analysis capable of being performed in most clinical laboratories. “We have made HSAFM analysis of the sample affordable by adapting common optical lasers and creating computer code to trace the DNA molecule,” said Dr Reed. “If commercialized, this process could offer a significantly cheaper alternative for detecting and diagnosing DNA length polymorphisms.”
He also emphasised that the method does not have any more complexity than a PCR assay and can easily be performed by most lab technicians.
As this technology will allow for more accurate and affordable diagnoses of genetic diseases, the next step for the VCU team is to test the technology in other settings where DNA structural mutations are relevant.
Profile:
Dr Jason Reed is associate professor in the Department of Physics at the Virginia Commonwealth University College of Humanities and Sciences in Richmond, Virginia, and member of the Cancer Biology research program at VCU Massey Cancer Center. His current research focuses on bio-mechanical systems, microfabrication and surface metrology for biological material characterization.
06.05.2021





