Long-standing mystery in membrane traffic solved
Researchers discovered that NSF unravels a single SNARE complex using one round ATP turnover by tearing the complex with a single burst, contradicting a previous theory that it unwinds in a processive manner.
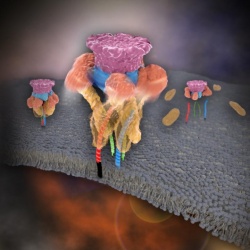
In 2013, James E. Rothman, Randy W. Schekman, and Thomas C. Südhof won the Nobel Prize in Physiology or Medicine for their discoveries of molecular machineries for vesicle trafficking, a major transport system in cells for maintaining cellular processes. Vesicle traffic acts as a kind of "home-delivery service" in cells. Vesicles package and deliver materials such as proteins and hormones from one cell organelle to another. Then it releases its contents by fusing with the target organelle's membrane. One example of vesicle traffic is in neuronal communications, where neurotransmitters are released from a neuron. Some of the key proteins for vesicle traffic discovered by the Nobel Prize winners were N-ethylmaleimide-sensitive factor (NSF), alpha-soluble NSF attachment protein (α-SNAP), and soluble SNAP receptors (SNAREs).
SNARE proteins are known as the minimal machinery for membrane fusion. To induce membrane fusion, the proteins combine to form a SNARE complex in a four helical bundle, and NSF and α-SNAP disassemble the SNARE complex for reuse. In particular, NSF can bind an energy source molecule, adenosine triphosphate (ATP), and the ATP-bound NSF develops internal tension via cleavage of ATP. This process is used to exert great force on SNARE complexes, eventually pulling them apart. However, although about 30 years have passed since the Nobel Prize winners' discovery, how NSF/α-SNAP disassembled the SNARE complex remained a mystery to scientists due to a lack in methodology.
In a recent issue of Science, published on March 27, 2015, a research team, led by Tae-Young Yoon of the Department of Physics at the Korea Advanced Institute of Science and Technology (KAIST) and Reinhard Jahn of the Department of Neurobiology of the Max-Planck-Institute for Biophysical Chemistry, reports that NSF/α-SNAP disassemble a single SNARE complex using various single-molecule biophysical methods that allow them to monitor and manipulate individual protein complexes.
"We have learned that NSF releases energy in a burst within 20 milliseconds to "tear" the SNARE complex apart in a one-step global unfolding reaction, which is immediately followed by the release of SNARE proteins," said Yoon.
Previously, it was believed that NSF disassembled a SNARE complex by unwinding it in a processive manner. Also, largely unexplained was how many cycles of ATP hydrolysis were required and how these cycles were connected to the disassembly of the SNARE complex.
Yoon added, "From our research, we found that NSF requires hydrolysis of ATPs that were already bound before it attached to the SNAREs--which means that only one round of an ATP turnover is sufficient for SNARE complex disassembly. Moreover, this is possible because NSF pulls a SNARE complex apart by building up the energy from individual ATPs and releasing it at once, yielding a "spring-loaded" mechanism."
NSF is a member of the ATPases associated with various cellular activities family (AAA+ ATPase), which is essential for many cellular functions such as DNA replication and protein degradation, membrane fusion, microtubule severing, peroxisome biogenesis, signal transduction, and the regulation of gene expression. This research has added valuable new insights and hints for studying AAA+ ATPase proteins, which are crucial for various living beings.
Source: Press release The Korea Advanced Institute of Science and Technology (KAIST)
30.03.2015