© Felix Petermann, MDC
News • AI approach to colorectal tumors
Deep learning identifies molecular patterns of cancer
An artificial intelligence platform developed at the Max Delbrück Center for Molecular Medicine (MDC) can analyze genomic data extremely quickly, picking out key patterns to classify different types of colorectal tumors and improve the drug discovery process. The deeper analysis shows some colorectal subtypes need to be reclassified.
A new deep-learning algorithm can quickly and accurately analyze several types of genomic data from colorectal tumors for more accurate classification, which could help improve diagnosis and related treatment options, according to new research published in the journal Life Science Alliance.
Colorectal tumors are extremely varied in how they develop, require different drugs and have very different survival rates. Often, they are classified into subtypes based on analysis of gene expression levels. “Disease is much more complex than just one gene,” said Altuna Akalin, bioinformatics scientist who leads the Bioinformatics Platform research group at MDC’s Berlin Institute of Medical Systems Biology (BIMSB). “To appreciate the complexity, we have to use some kind of machine learning to really make use of all the data.”
To look at numerous features contained in genetic material, including gene expression, single point mutations and DNA copy-numbers, Akalin and PhD student Jonathan Ronen designed the Multi-omics Autoencoder Integration platform – “maui” for short.
Supervised machine learning typically requires human experts to label data and then train an algorithm to predict those labels. For example, to predict eye color from pictures of eyes, the researchers first feed the algorithm with pictures where eye color is labeled. The algorithm learns to identify different eye colors and can independently analyze new data.
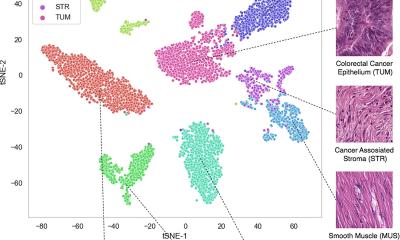
In contrast, unsupervised machine learning does not involve training. A deep-learning algorithm is fed data without labels and sifts through it to find common patterns or representative features, which are called latent factors. For example, this kind of algorithm can process pictures of faces that are not labeled in any way, then identify key features, like eye colors, eyebrow shapes, nose shapes, smiles. As a deep-learning platform, maui is able to analyze multiple “omics” datasets and identify the most relevant patterns or features, in this case, gene sets or pathways to colorectal cancer.
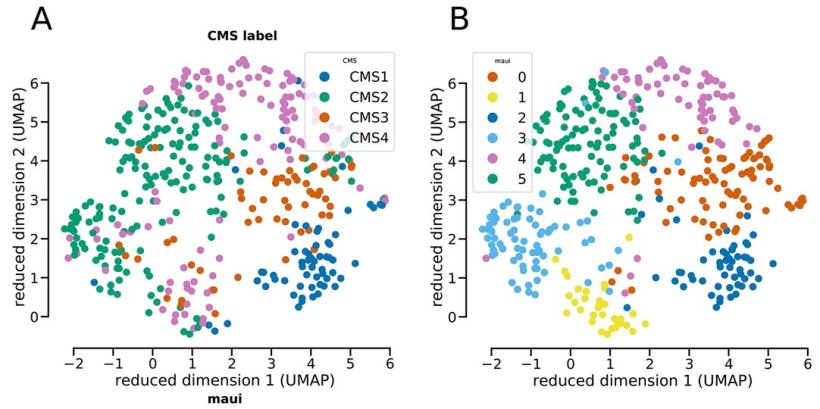
© Akalin Lab, MDC
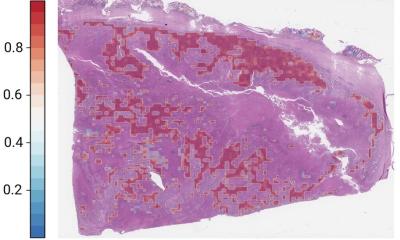
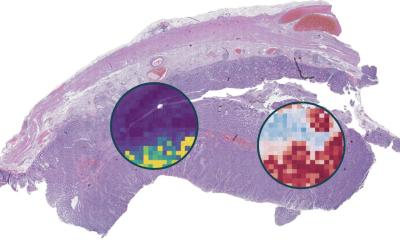
maui identified patterns associated with the four established subtypes of colorectal cancer, assigning tumors to subtypes with high accuracy. It also made an interesting discovery. The platform found a pattern that suggests one subtype (CMS2) might need to be split into two separate groups. The tumors have different mechanisms and survival rates. The team suggests further investigation to verify if the subtype is unique or perhaps representative of the tumor spreading. Still, it demonstrates the power of the platform to take all the data, rather than only the known genes associated with a disease, and produce deeper insights. “Data science can handle complex data that is hard to handle other ways and makes sense of it,” Akalin said. “You can feed it everything you have on the tumors and it finds meaningful patterns.”
It is able to learn orders of magnitude more latent factors, at a fraction of the computation time
Jonathan Ronen
The program was not just more accurate, it also works much faster than other machine learning algorithms – three minutes to pick out 100 patterns, compared to the other programs that took 20 minutes and 11 hours. “It is able to learn orders of magnitude more latent factors, at a fraction of the computation time,” said Jonathan Ronen, first author of the research.
The team was actually surprised at how fast the system performs, especially because they did not have to use graphics cards that usually help speed up calculations. This shows how extremely well optimized, or efficient, the algorithm is, though they are continuing to find ways to further finetune the system.
The team, which also included Bayer AG computational biologist Sikander Hayat, adapted their program to analyze cell lines taken from tumors and grown in labs for researching the effects of potential drug treatments. However, cell lines differ from real tumors in many ways on the molecular level. The team used maui to compare cell lines currently used for testing colorectal cancer drugs to see how closely they were related to real tumors. Nearly half of the lines were found to be more related to other cell lines than actual tumors. A handful were found to be the best lines most closely representing the different classes of CRC tumors.
While drug discovery research is moving away from cell lines, this insight could help maximize the potential impact of cell line research, and could be adapted for other types of genetic-based drug testing tools.
Now that the deep-learning platform for colorectal cancer has been established, it could be used to analyze data for new patients. “Think of this like a search engine,” Akalin said. A clinician could input the new patient’s genetic data into maui to find the closest match to quickly and accurately classify the tumor. The platform could advise what drugs have been used on the closest matching tumors and how well they worked, thus helping to predict drug responses and survival outlook.
For now, this could take place in a research setting only after doctors have tried the established protocols. It is a long road for a test or system to be approved for clinical use, Akalin said. The team is exploring the potential for commercialization with the help of the Berlin Institute of Health’s Digital Health Accelerator Program. They are also in the process of adapting maui for other types of cancers.
Source: Max Delbrück Center for Molecular Medicine
13.06.2022