Article • Human-bacterial symbiosis
The value of the human microbiome
The human species maintains symbiotic relationships with a multitude of microbial organisms that colonise the inside as well as the surface of the body. Scientists, for a long time, underestimated the significance of these organisms for humans.
Report: Sandra Theison
By far the most bacteria of the microbiome can be found in the gastrointestinal tract, particularly in the colon. The number of microbes in the body is estimated to exceed ten times the number of human cells; the body thus contains 100-fold more unique genes than the human genome. Only 10 to 30 percent of these microbial organisms are culturable. In humans, most bacteria of the gastrointestinal microbiome belong to the four phyla Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria.
In clinical care, the identification of bacteria is crucial to be able to distinguish between bacteria that are colonisers or those that originally cause an infection. Today, this identification is achieved via sequencing of 16S rRNA, particularly if phenotypic methods of bacterial identification are difficult.
The bacterial 16S rRNA gene codes for the subunit 30S of the prokaryotic ribosome 70S and is thus present in the genome of all bacteria. Since it differs from its eukaryotic homologue 18S rRNA, precise identification of bacterial DNA versus human DNA is possible. The gene sequence of this subunit is used in evolution biology as the instrument for phylogenetic analysis.
The 16S rRNA gene consists of approx. 1,500 base pairs and can be separated into several conserved and nine less conserved regions (V1-V9). Since the less conserved regions have little impact on the formation of the subunit 30S, they are hypervariable and specific to particular bacterial species. These regions may thus be used as taxonomic markers. During a polymerase chain reaction (PCR), primers can dock onto the known conserved areas and selectively amplify the variable regions. Sequencing of these DNA sections is usually done by pyrosequencing (see box below).
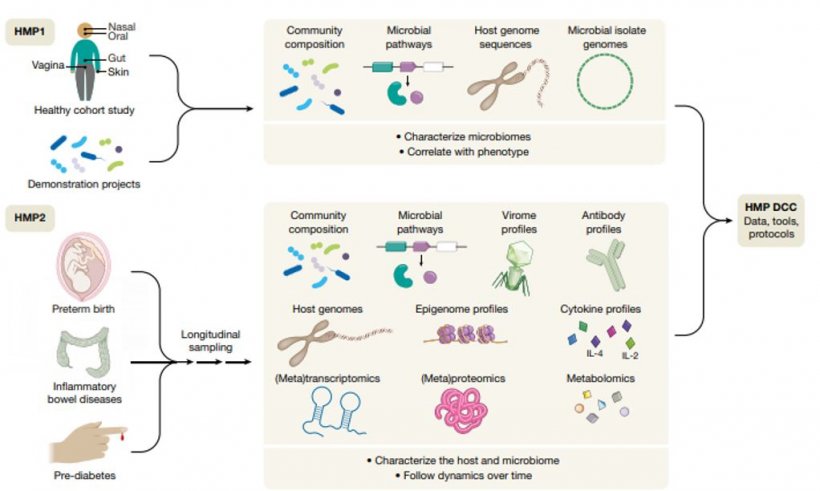
The Human Microbiome Project
The Human Microbiome Project (HMP) is one of the largest biological projects ever. It aims to identify all microbes that colonise the human body and study their roles in health and disease states. HMP was launched in 2008 by the US National Institute of Health (NIH), focusing on five body sites of a healthy individual: skin, nose, mouth, gut and vagina. In 2013, HMP1 was completed after having collected 14 TB of data from the samples of 300 individuals.
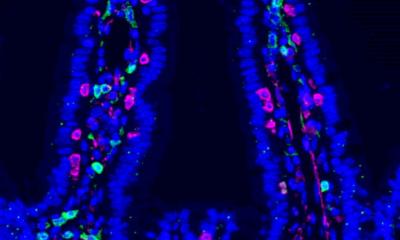
The second phase of HMP, also called Integrative HMP (HMP2/iHMP), was launched in 2014. HMP2 was designed to explore host-microbiome interplay and focused on three topics: pregnancy & pre-term birth, inflammatory bowel diseases (IBD) and onset of Type 2 diabetes (T2D). By way of example, the procedure regarding pre-term birth is described in the figure shown above.
There are significant disparities in the frequency of pre-term birth. In the United States, for example, American women of African ancestry are at a much higher risk of pre-term birth than any other group of women. 16S rRNA analysis was performed and additional profiles from 45 pre-term births and 90 term births were established. Early on in the selection process, it became clear that the percentage of pre-term birth is higher among women of African ancestry. The analyses showed that women who delivered pre-term exhibited significantly lower vaginal levels of Lactobacillus crispatus and higher levels of 13 other bacterial taxa. Pre-term-birth-associated taxa were correlated with proinflammatory cytokines in vaginal fluid. These findings highlight new opportunities for assessment of the risk of pre-term birth.
The way forward
Future research might focus on identifying and characterising the microbiome with regard to itproteome and metabolome and their interaction with human proteins and metabolites.
Pyrosequencing
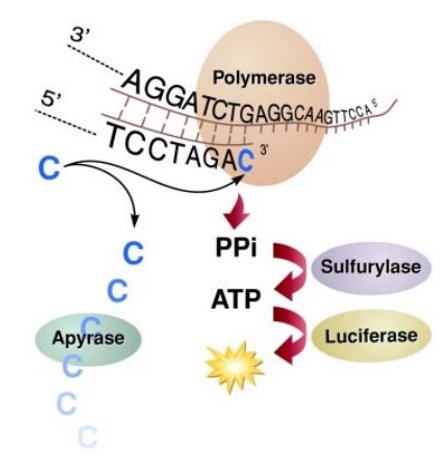
As in Sanger sequencing, the DNA polymerase originating in a primer elongates the complementary DNA strand. When DNA polymerase adds a complementary nucleotide to the DNA strand to be synthesised, pyrophosphate is released. The enzyme ATP sulfurylase converts the pyrophosphate into adenosintriphosphate, which triggers the luciferase reaction where luciferine is turned into oxyluciferine. The intensity of light emission during this process is proportional to the amount of adenosintriphosphate used. Unincorporated nucleotides are degraded by apyrase. Since the individual nucleotides are sequentially added to the reaction, the temporal pattern of light emission allows determination of the DNA sequences.
13.07.2020