Cancers
Computer program uncovers hidden genomic alterations
Cancer is rarely the result of a single mutation in a single gene. Rather, tumors arise from the complex interplay between any number of mutually exclusive abnormal changes in the genome, the combinations of which can be unique to each individual patient. To better characterize the functional context of genomic variations in cancer, researchers at University of California San Diego School of Medicine and the Broad Institute developed a new computer algorithm they call REVEALER.
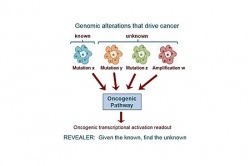
This tool is designed to help researchers identify groups of genetic variations that together associate with a particular way cancer cells get activated, or how they respond to certain treatments. REVEALER is available for free to the global scientific community via the bioinformatics software portal GenePattern.org
“This computational analysis method effectively uncovers the functional context of genomic alterations, such as gene mutations, amplifications, or deletions, that drive tumor formation,” said senior author Pablo Tamayo, PhD, professor and co-director of the UC San Diego Moores Cancer Center Genomics and Computational Biology Shared Resource.
Tamayo and team tested REVEALER using The Cancer Genome Atlas (TCGA), the National Institutes of Health’s database of genomic information from more than 500 human tumors representing many cancer types. REVEALER revealed gene alterations associated with the activation of several cellular processes known to play a role in tumor development and response to certain drugs. Some of these gene mutations were already known, but others were new. For example, the researchers discovered new activating genomic abnormalities for beta-catenin, a cancer-promoting protein, and for the oxidative stress response that some cancers hijack to increase their viability.
REVEALER is a powerful approach but requires as input high-quality genomic data and a significant number of cancer samples, which can be a challenge, Tamayo says. But REVEALER is more sensitive at detecting similarities between different types of genomic features and less dependent on simplifying statistical assumptions, compared to other methods, he says.
“This study demonstrates the potential of combining functional profiling of cells with the characterizations of cancer genomes via next generation sequencing,” said co-senior author Jill P. Mesirov, PhD, professor and associate vice chancellor for computational health sciences at UC San Diego School of Medicine.
Source: UC San Diego School of Medicine
27.04.2016