Image source: Randhawa et al., PLOS ONE 2020 (CC BY 4.0)
News • Coronavirus origins
Researchers crack COVID-19 genome signature
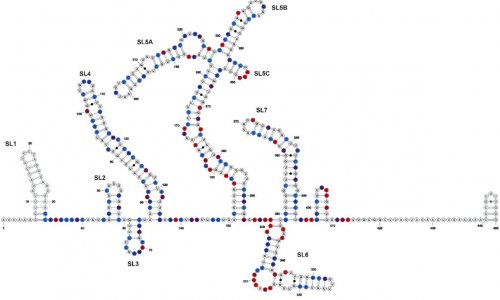
Using machine learning, a team of Western computer scientists and biologists have identified an underlying genomic signature for 29 different COVID-19 DNA sequences.
This new data discovery tool will allow researchers to quickly and easily classify a deadly virus like COVID-19 in just minutes – a process and pace of high importance for strategic planning and mobilizing medical needs during a pandemic. The study also supports the scientific hypothesis that COVID-19 (SARS-CoV-2) has its origin in bats as Sarbecovirus, a subgroup of Betacoronavirus.
The findings, Machine learning using intrinsic genomic signatures for rapid classification of novel pathogens: COVID-19 case study, were published in PLOS ONE.
All we needed was the COVID-19 DNA sequence to discover its own intrinsic sequence pattern
Kathleen Hill
The “ultra-fast, scalable, and highly accurate” classification system uses a new graphic-based, specialized software and decision-tree approach to illustrate the classification and arrive at a best choice out of all possible outcomes. The entire method uses a new graphic-based, specialized software to illustrate a best choice out of all tested possible outcomes. Biology professor Kathleen Hill co-led the study with Western collaborators in Computer Science and Statistical and Actuarial Sciences, along with others in the University of Waterloo’s Department of Computer Science.
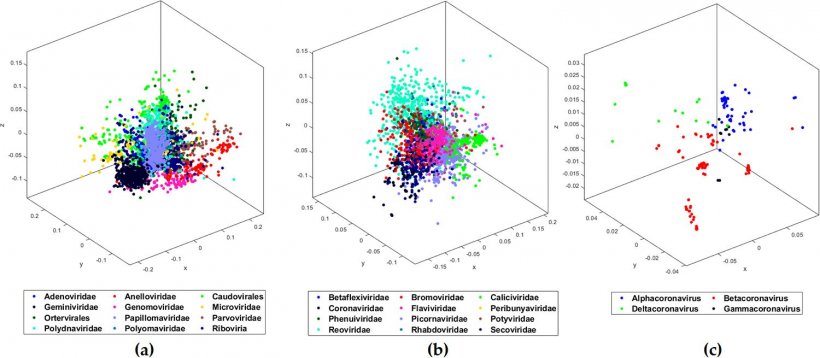
The machine-learning method achieves 100 per cent accurate classification of the COVID-19 sequences and more importantly, discovers the most relevant relationships among more than 5,000 viral genomes again within minutes. “All we needed was the COVID-19 DNA sequence to discover its own intrinsic sequence pattern. We used that signature pattern and a logical approach to match that pattern as close as possible to other viruses and achieved a fine level of classification in minutes – not days, not hours but minutes,” Hill said.
This classification tool has already been used to analyze more than 5,000 unique viral genomic sequences, including the 29 COVID-19 sequences available on Jan. 27. Hill believes the tool, which is able to classify any newly discovered virus sequence COVID-19 or otherwise, will be an essential component in the toolkit for vaccine and drug developers, front-line health-care workers, researchers and scientists during this global pandemic and beyond.
Source: Western University
04.05.2020