Deep Learning
Deep Learning predicts hematopoietic stem cell development
Autonomous driving, automatic speech recognition, and the game Go: Deep Learning is generating more and more public awareness. Scientists at the Helmholtz Zentrum München and their partners at ETH Zurich and the Technical University of Munich (TUM) have now used it to determine the development of hematopoietic stem cells in advance. In ‘Nature Methods’ they describe how their software predicts the future cell type based on microscopy images.
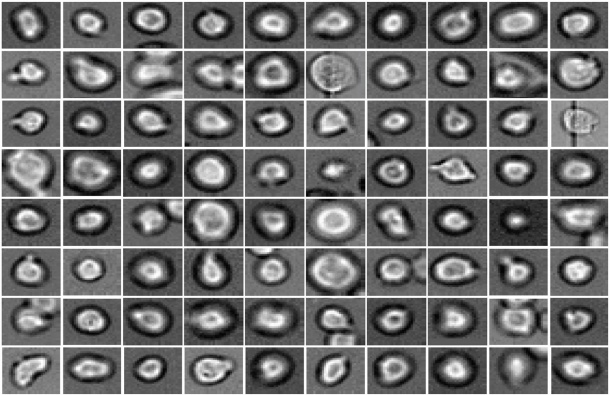
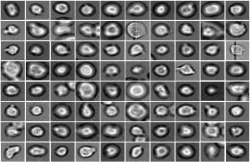
Today, cell biology is no longer limited to static states but also attempts to understand the dynamic development of cell populations. One example is the generation of different types of blood cells from their precursors, the hematopoietic stem cells. “A hematopoietic stem cell's decision to become a certain cell type cannot be observed. At this time, it is only possible to verify the decision retrospectively with cell surface markers,” explains Dr. Carsten Marr, head of the Quantitative Single Cell Dynamics Research Group at the Helmholtz Zentrum München's Institute of Computational Biology (ICB).
He and his team have now developed an algorithm that can predict the decision in advance. So-called Deep Learning is the key. “Deep Neural Networks play a major role in our method,” says Marr. “Our algorithm classifies light microscopic images and videos of individual cells by comparing these data with past experience from the development of such cells. In this way, the algorithm ‘learns’ how certain cells behave.”
Three generations earlier than standard methods
Specifically, the researchers examined hematopoietic stem cells that were filmed under the microscope in the lab of Timm Schroeder at ETH Zurich.* Using the information on appearance and speed, the software was able to ‘memorize’ the corresponding behaviour patterns and then make its prediction. “Compared to conventional methods, such as fluorescent antibodies against certain surface proteins, we know how the cells will decide three cell generations earlier,” reports ICB scientist Dr. Felix Buggenthin, joint first author of the study together with Dr. Florian Büttner.
But what is the benefit of this look into the future? As study leader Marr explains, “Since we now know which cells will develop in which way, we can isolate them earlier than before and examine how they differ at a molecular level. We want to use this information to understand how the choices are made for particular developmental traits.”
In the future, the focus will expand beyond hematopoietic stem cells. “We are using Deep Learning for very different problems with sufficiently large data records,” explains Prof. Dr. Dr. Fabian Theis, ICB director and holder of the Mathematical Modelling of Biological Systems Chair at the TUM, who led the study together with Carsten Marr. “For example, we use very similar algorithms to analyse disease-associated patterns in the genome and identify biomarkers in clinical cell screens.”
Further Information
* The study described here is the latest result of a close cooperation between the ICB scientists and Prof. Dr. Timm Schroeder from the Department of Biosystems Science and Engineering at ETH Zurich in Basel, who previously worked at the Helmholtz Zentrum in Munich. In July 2016, the scientists jointly introduced a software in ‘Nature Biotechnolgy’ that allows to observe individual cells over many days and simultaneously measure their molecular properties. https://www.helmholtz-muenchen.de They also published a study in ‘Nature’ that already dealt with the development of hematopoietic stem cells. Using time-lapse microscopy, the researchers were able to observe the maturation of living hematopoietic stem cells with high precision while also quantifying certain proteins. https://www.helmholtz-muenchen.de/
Source: Helmholtz Zentrum München - German Research Center for Environmental Health
23.02.2017