Image source: Nunes L et al., Nature 2024 (CC BY 4.0)
News • Genome and transcriptome analysis
Colorectal cancer prognosis derived from genetic signatures
Using a unique collection of genetic and clinical data for colorectal cancer, researchers at Uppsala University have revealed genetic new alterations and developed of a new molecular classifier of tumour variants.
The finding could lead to improved possibilities for individualised therapies. This is shown in study recently published in the journal Nature.
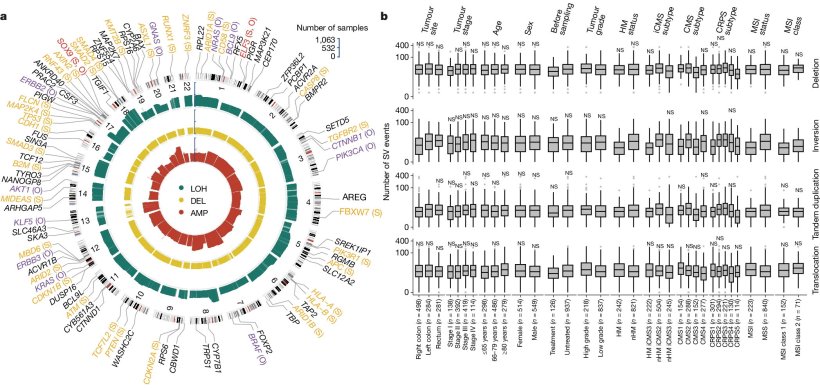
Colorectal cancer is the third most common and the second deadliest tumour type. The disease is caused by mutations in cancer driver genes that result in un-controlled cell growth. To understand the link between the genetic alterations and disease development and prognosis, the Uppsala researchers and their collaborators have sequenced the DNA from tumour and normal tissues from more than one thousand colorectal cancer patients.
“Since we also have access to clinical information from these patients, our work has resulted in one of the world’s largest colorectal cancer cohorts with comprehensive clinical and genomic data. The scope of this dataset is ground-breaking and offers great amounts of information for scientists in the area, says Tobias Sjöblom, professor at the Department of Immunology, Genetics and Pathology and one of the researchers who led the study.

Image source: Uppsala University; photo: Mikael Wallerstedt
In the first publication from the cohort, the researchers have identified almost one hundred mutated cancer driver genes. Out of these, 9 were novel to colorectal cancer, and 24 to any cancer. The researcher also examined what kinds of genetic alteration that were present, including small mutations and larger structural variations, and alterations that affected gene activity.
“The genetic information that we obtained was then compared with clinical information, particularly survival data: This way we identified key prognostic genomic alterations in specific patient groups. Some of these findings confirm previous research, while others are novel discoveries, says Luis Nunes, researcher at the Department of Immunology, Genetics and Pathology at Uppsala University and one of the first authors of the study.
The scientists have also used the results from the study to develop a new strategy to molecularly classify colorectal cancer. “Our classifier is based on gene activity and with this we could effectively categorise the majority of tumours into five prognostic subtypes with distinct molecular features. This refines and adds into previous colorectal cancer subtyping classifiers and provides a clearer prognostic picture of different groups of tumours, says Luis Nunes.
The scientists behind the study believe that their discovery of prognostic mutations and new molecular subtypes of tumours are important both for future studies of colorectal cancer genes and for the development of innovative diagnostic and therapeutic strategies. “Our study marks a significant step forward in the fight against colorectal cancer. We hope that it will bring us closer to precision medicine with more effective treatments and improved patient outcomes,” says Tobias Sjöblom.
Source: Uppsala University
09.08.2024