DNA
Blood test may expand scope of liquid biopsies
When cells die, they don't vanish without a trace. Instead, they leave behind their fingerprints in the form of cell-free DNA. In people, these tiny fragments of DNA can be found in the bloodstream.
In recent years, research into cell-free DNA has led to a type of test, dubbed the "liquid biopsy," that can diagnose and monitor some cancers, identify fetal abnormalities, and assess the health of transplanted organs, all with a simple blood draw. Despite the promise of these tests, at present the range of conditions they can detect is limited.
In a new study, UW Medicine researchers in Seattle show that it might be possible to overcome these limitations with a method for identifying what types of cells gave rise to the cell-free DNA.
This method could potentially expand the scope of the liquid biopsy. The new approach relies on analyzing the fragmentation patterns seen in the cell-free DNA in one individual, and comparing that pattern to what might be expected for cell death associated with various medical or physiological conditions.
The research was led by Matthew W. Snyder, a graduate student, and Dr. Martin Kircher, a postdoctoral fellow, in the laboratory of Dr. Jay Shendure, University of Washington professor of genome sciences and a Howard Hughes Medical Institute investigator.
Their results are reported this week in the scientific journal Cell. Their paper is titled, "Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin."
"Our findings suggest it is possible to identify tissues contributing to cell-free DNA by looking at these fragmentation patterns, instead of looking for specific mutations in the DNA," Shendure said. The test examines the ends of each fragment of DNA, and tries to identify hotspots, or parts of the DNA that get cut more frequently than others.
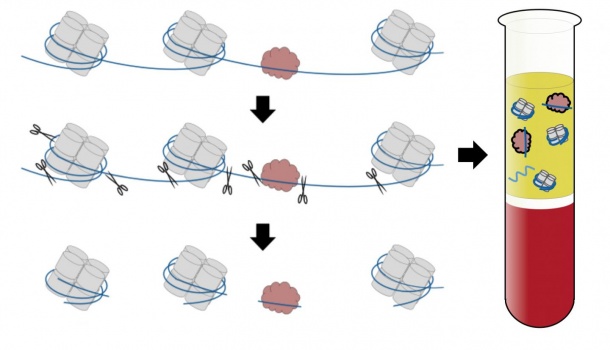
To fit into a cell's nucleus, DNA has to be coiled, wound and folded into an incredibly compact package. Key to this process are structures called nucleosomes, which are composed of a protein core around which the DNA is wrapped like thread around a spool. Nucleosomes are strung out along the entire length of the genome, like beads on a string, with the DNA looping around one nucleosome after the next. Each cell type in the body packages DNA slightly differently. These differences leave their tell-tale mark in the resulting cell-free DNA.
The UW researchers hypothesized that they could use these fingerprints to work backwards to try to figure out where the cell-free DNA originated. To do this, they first needed to know which parts of the DNA were wrapped around the nucleosomes.
During cell death, the DNA gets chopped into small pieces by enzymes that like to cut in the easily accessible, unprotected sections of DNA in between the nucleosomes. Research has pointed to 13 million positions in the human genome where nucleosomes are most likely to be located.
Using blood samples from cancer patients, the researchers showed that different types of cancer left different nucleosome fingerprints in the cell-free DNA. For some of the cancers, the researchers could identify the anatomical source of the tumor.
"This could be particularly relevant in the 5 percent of metastatic cancers whose original source is unknown," Shendure said, adding that the test "could aid in diagnosing what kind of cancer it is and to help guide treatment."
Most liquid biopsy approaches look for specific DNA mismatches between different cells in the body, such as the mutations found in tumor cells but not healthy cells. The researchers think that the advantage of the new test is that it may work even when the cells are genetically identical to one another.
The approach potentially could be used to diagnose a wide variety of conditions that kill off cells, but that as yet cannot be diagnosed with liquid biopsies. Examples of such serious, cell-destroying medical problems are heart attacks, strokes and autoimmune diseases.
Source: University of Washington Health Sciences/UW Medicine
15.01.2016