Linac Coherent Light Source
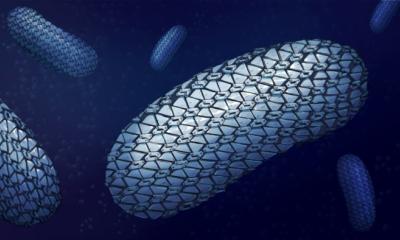
A glimpse into live bacteria
A group of scientists in Sweden has taken an important step towards the goal of peering inside a working cell. They are among researchers around the globe who are seeking a method that enables the observation of proteins, lipids and DNA inside individual cells, as well as gaining a better understanding of how this intricate and interconnected system changes with time.
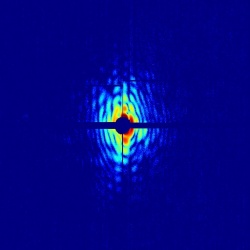
The scientists from Sweden's Uppsala University have developed a container-free injection method to introduce a beam of live bacteria into the micron-sized focus of the Linac Coherent Light Source (LCLS) at the SLAC National Accelerator Laboratory in Menlo Park, Calif. The LCLS is "like a high-speed camera with an incredibly bright flash" and can take X-ray snapshots that freeze chemical or biological reactions in time, according to the SLAC website.
Gijs van der Schot, a Ph.D. student in the Department of Cell and Molecular Biology at Uppsala said: "The X-ray laser we use for our work, the LCLS, is a fascinating machine -- because of the physical principles behind it and the precise engineering of its parts -- that produces very bright and ultra-short pulses."
In the machine, an ultrafast pulse of X-ray light is fired at a target. In the Swedish researcher's case, the target was a beam of live blue-green algae, called cyanobacteria. When the light hits the cells, the waves scatter and interfere with each other, forming a pattern of light called a diffraction pattern. "The structure of the cell is hidden inside the wavy diffraction pattern we measure," van der Schot explained. The team used a mix of mathematical techniques to retrieve the best-fitting two-dimensional reconstruction of the cell.
To make the reconstruction imaging methods work, the light must be coherent, meaning that all the electromagnetic waves are in sync, and the pulses must be very bright. So bright, in fact, it appears that all the light hitting Earth is confined to a square millimeter. "The length of the light pulses must also be ultra-short to outpace the damaging processes," van der Schot noted. In other words, the light pulse has to travel all the way through the cell before the cell starts to react to the damaging X-rays.
Another important aspect of the Swedish group's work is "an injector that introduces cells into the X-ray beam," van der Schot added. "It produces the beam of aerosolized live cyanobacteria with very little solvent, which allows for low-noise diffraction patterns. Combined with the 120-Hertz repetition rate of the X-ray laser, we can obtain more than a million 2-D images of cells daily." The significance of the team's approach is that it "enables imaging of live cells in a high-throughput manner at a time resolution at which all biologically relevant dynamics appear static," he said.
Earlier work by the group showed the theoretical possibility of reaching sub-nanometer spatial resolution. Their latest work has encountered no fundamental limitations so far, and the results are as they had predicted. "We measured the diffraction signal up to 4-nm [nanometer] resolution -- the size of a small protein -- and produced 2-D images at 76-nm resolution," said van der Schot. "The main limitation of our resolution is the restricted dynamic range of the detector. This means that the most intense images -- reaching 4-nm resolution scattering -- are overexposed in the center of the diffraction pattern, which renders image reconstruction impossible."
Because of that limitation, the researchers developed a filter that significantly improves the dynamic range and are now "prepared to measure again," van der Schot said. "With the new technologies we've developed, we should be able to create 2-D models of cells to within 1- to 10-nm resolution by next year." The team published their first images of live bacteria in the journal Nature Communications in February. They continue to work to improve the process.
The next step is to go three-dimensional (3-D). "Since the technique allows only one shot per particle, it's no trivial matter to combine multiple 3-D patterns to create one 3-D model. All cells are different," van der Schot pointed out. "Millions of 2-D patterns must represent more than 2-D, however. We can use the larger number of 2-D images to derive semi-3-D information about cellular processes such as division and organelle formation." He estimates that it will take an extra year or two to develop the methods to enable this advance.
As for future applications, the group's method will offer researchers a new way to study processes such as cell division and virus infection. "The large number of images allow us to examine the processes from many angles and points in time," van der Schot said.
Source: Sweden's Uppsala University
22.10.2015