Article • Digital pathology
Boosting diagnostic accuracy and efficiency
Certain members of Generation Y, who grew up alongside enormous information technology (IT) advances, now occupy decision-making roles. Meanwhile, generation Z is emerging into the continuing IT revolution; the digital world surrounds us.
Information spreads and is accessed worldwide, unlimited and exploited in all life’s sectors, including the most complex digitisation of health service administration and diagnostic services – particularly histopathology.
Due to its complexity and huge size of digital slides, histopathology requires the harmonisation of professional and medico-legal aspects with IT and process automation efforts at high standards, to help clinicians to offer patients the best accessible cure in time.
Microscopic study of stained tissue sections at thickness of ~1/300th of a millimetre, enables surgical pathologists to reveal structural and molecular alterations in disease processes to determine diagnostic entities and their biological behaviour. Their diagnoses are essential to clinically manage patients properly. The integrative role of pathologists, particularly in specifying malignant tumours and screening for dynamically growing number of biomarkers related to patients’ response to molecular targeted therapy, upgrades their responsibility in patient recovery rates.
Recent progress in molecular technology has allowed better understanding of the pathobiology of disease processes, which need to be correlated with tissue and cell morphology to understand structure-function relationships. Techniques of ‘molecular morphology’ which combine structural and molecular information including genomic in situ hybridisation (ISH) to detect chromosome and gene abnormalities, and immunohistochemistry (IHC) to reveal translated proteins determining normal and abnormal cell functions, are increasingly important.
In tumour pathology, series of biomarkers need to be tested semi-quantitatively, at the cellular or even subcellular level within the tissue architecture. The amount and complexity of information to be accessed and handled by pathologists require support from artificial intelligence of computing power. Hospital administration and diagnostic reporting systems already exploit digital data management, which integrate alphanumerical data, still digital images of macroscopic samples and radiology images, as well as voice recognition.
By now, advanced slide scanners allow digitisation of whole microscopic slides at high resolution and colour fidelity, which can also be integrated into the digital diagnostic workflow. Despite the enormous sizes of digital slides measured in gigabytes, IT management systems, including software tools and computer networks, can now support their sharing and exploitation for teaching, consultation, quality assurance, research and diagnostic activities in histopathology.
The digital side
Digital slide scanners create the computerised representation of whole microscopic slides with all crucial details of stained tissue sections of 3-5 micrometre thickness, which then can be viewed remotely, if uploaded to a server computer.
New generation panoramic area scanners use motorised stage to digitise microscopic fields of view (FOVs) at enormous speed, exceeding 150 FOVs per second, supported by a synchronised flashing light source.
During scanning, a stitching software algorithm makes FOV tile borders smooth and seamless. Advanced systems combine brightfield scanning to emulate transmission microscopy, and fluorescence scanning supported with up to nine emission filters to substitute the multichannel epifluorescence microscope.
Photons emitted by the sample are collected by cameras of high-density complementary metal oxide semiconductor (CMOS) sensors and translated into digital signals.
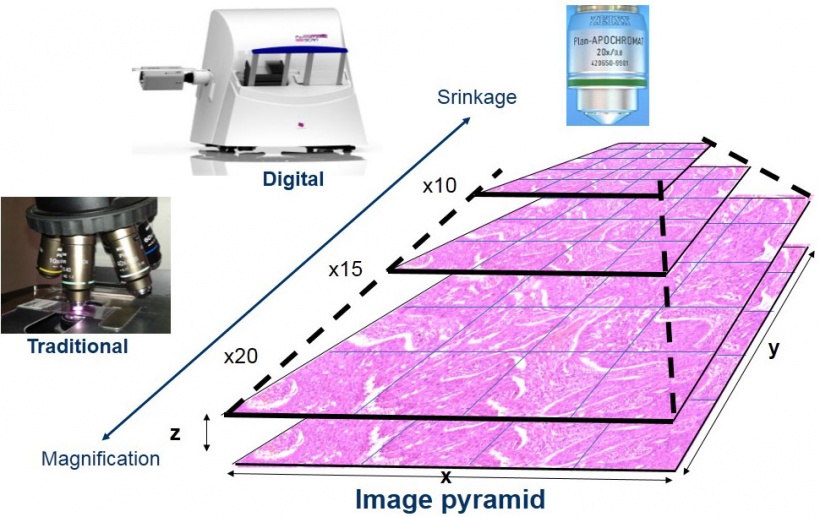
High spatial resolution is achieved using x20 microscope objectives with the highest available numerical aperture (NA=0.8), while NA=0.9, x40 objectives offer somewhat better resolution for cytology samples. Image sharpness and high colour fidelity is ensured by automatic focusing and white balance. Digital slides are built up as pyramids of microscopic image series made up of pixel arrays organised along the x-y coordinates, where low and high power views are generated by compressing or magnifying the original sharp and optimally lit images (Figure 1).
As with Google Earth maps, which allow one to rapidly navigate through cities at low power to find streets or buildings by zooming into the image, the Viewer software enables pathologists to test, for example, gastric biopsies and zoom into glands for analysing their cell components and microenvironment to see if they are abnormal and to what extent.
Digital microscopy adds unlimited computer technology potential to traditional microscopy
Digital microscopy offers functions that are not available for an optical microscope. Viewing slides through the computer monitor, with easy access to the computer’s multi-functionality, is much more ergonomic than peeping through the narrow ocular of an optical microscope. Even pathologists with great affection for conventional microscopy acknowledge this, after spending enough time using this new technology.
Dedicated software tools provide unlimited potential to extract all inherent information from digital slides and integrate results into diagnostic reports and hospital databases.
The viewer interface utilises the whole computer monitor and the mouse to select, drag, move and zoom into any image details where preview images and navigation history of high power analysis are also simultaneously traced. Scanning through several focus levels within the whole sample thickness offers access to the z-dimension used to emulate fine focusing of the optical microscope. This function, which can be crucial when testing small (<1 micrometre diameter) objects or signals, allows in-focus photo-documentation of all important details, when merging sharp image layers into one extended focus, to overcome the focal depth limitation of high resolution objectives. Multilayer digitisation of FISH signals, which are small (<1 mm) and fade quickly, offers permanent samples for clear signal recognition and analysis without temporal limitations.
In-focus navigation at any arbitrary magnification without the need to change objectives, realign the focus or adjust the lighting conditions can facilitate pathology workflow. Automated alignment into the same orientation of serial sections offers further support. Additionally, several slides, even of bright-field and fluorescence, can be viewed side-by-side on the same monitor and linked for comparative analysis.
Image pixels of calibrated dimensions allow straight measurements of object distance, perimeter and area, while image segmentation along distinct object size, colour and intensity permits automated object quantification by automated image analysis algorithms. Permanent annotations and text put on digital slides and still-image archiving of publication quality are also exclusive features of digital microscopy. For instance, clinical pathology meetings and onco-team consultations are greatly facilitated by promptly taken still images of any important area at any magnification.
One of the most significant benefits of digital slides is their share ability through the intranet or internet with an unlimited number of partners. Furthermore, compared to physical section archiving the logistics of digital slide storage, sorting and access become simple tasks managed with a computer for interpretation into diagnostic reports and guiding clinicians to find the most relevant and effective therapy.
Digital pathology: digital slides-based diagnostics
By now, digital scanning technology, the software tools for digital slide access and handling, as well as barcoding identification of samples and IT support, including computer networks and hospital databases, have matured into the stage that makes digital microscopy an attractive option for primary diagnostics. High resolution and colour fidelity, permanently in-focus digital slides meet diagnostic requirements particularly when colour calibrated high resolution monitors are used for user-friendly interface.
Slide control is supported by a professional joystick driver imitating all functions of the macro-, micrometre and stage guiding knobs of a traditional microscope. Advanced digital workstations utilise shared double monitors, one to access and arrange information on patient history from the hospital database system (HIS) including imaging diagnostic data, earlier pathology reports and treatment data, and another for current diagnostic reporting with macroscopic descriptions and photos, digital slide series and a reporting area.
To manage such a complex array of information, including alphanumerical and imaging data, dedicated software tools are also available. The CaseCenter software supports access and integration of digital slides into the reporting system. CaseViewer is specialised for convenient analysis of digital slides, including parallel slide views, and prompt, automated arrangement of serial slides involving those stained for immunohistochemistry or in situ hybridisation into the same orientation. This allows easy selection of interesting areas, either for further empirical or for automated image segmentation-based biomarker assessment.
Validated image analysis algorithms are available to accelerate efficiency and prevent subjective bias, particularly in assessing biomarker expression linked to molecular targeted therapies.
Reliable automated testing of genomic abnormalities using image analysis of FISH signals is also an attractive option today.
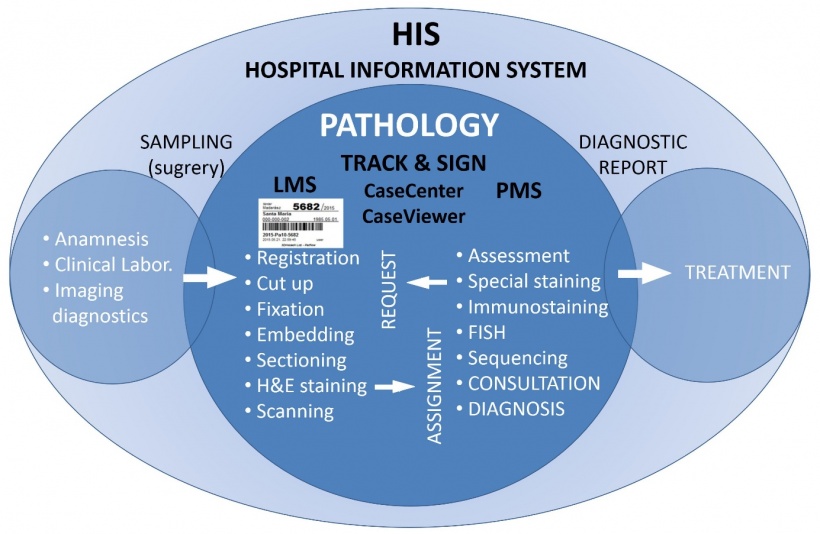
Pathology is based on examining and classifying surgically removed tissues, cytology or blood samples into diagnostic entities. The quality of sample management and pre-analytical processing fundamentally determines the success of both the analytical (staining tests) and post-analytical (interpretation) results. Therefore, sample tracing for controlling appropriate pre-analytical conditions is an equally important requirements in digitisation of the whole pathology workflow.
The ‘Track & Sign’ software environment supports barcode-based sample tracking from its arrival up to signing up the diagnostic report. It allows checking the status of any sample and its progeny (tissue, cell specimen, slide, cytospin, protein or nucleic acid isolates etc.) during the whole pathology workflow by reading sample barcodes at workstations the given sample passes through. These steps include fixation, embedding, cutting, staining and archiving in digital slide server repositories (Figure 2).
The software package also controls the diagnostic reporting workflow by indicating the stages of examination of stained slides as ‘new’, ‘under examination’ or ‘closed’, and allows initiating new stains or molecular pathology requests. Therefore, the Track & Sign system serves as a pathology information system (PIS) that can communicate with any database-supported HIS system.
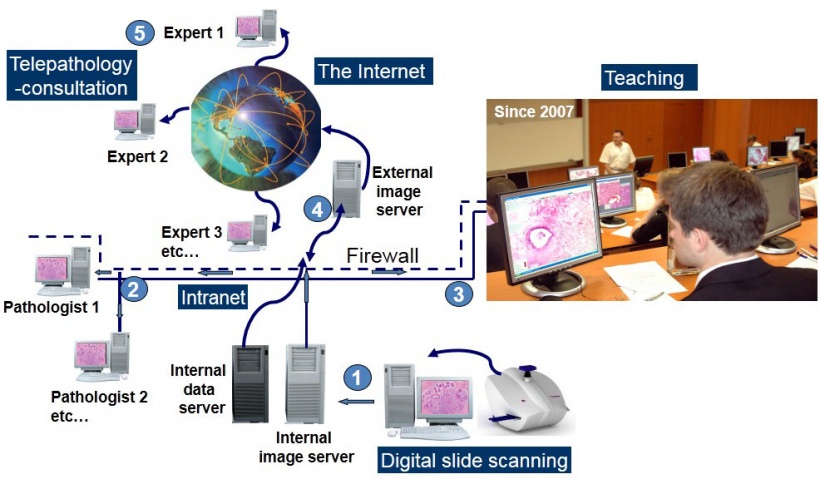
Interactive teaching and dynamic telepathology
Based on using digital slides, online teaching and telepathology (abbreviation of telecommunication pathology) require very similar informatics support and can be executed either through intranet or the internet (Figure 3).
Digital teaching and continuous education of anatomical pathology are justified both by versatility and reasonable costs. When one considers the burden of purchasing and maintaining optical microscopes and the regular replacement of tutorial glass slides, cost saving is obvious. A great benefit of digital microscopy for teaching lies in the access by everyone to the best selection of standard slides without spatial or temporal limitations.
Furthermore, a single digital slide can demonstrate a rare entity or any expensive ancillary technique, such as immunohistochemistry or FISH, by serving unlimited number of students. Additional rewards include parallel viewing of two or more slides or opening e.g. several copies of the same slide and correlating distant areas at high power simultaneously side-by-side to demonstrate tumour heterogeneity.
Annotations and text inserted into specified regions of slides help guide students at distant learning and self-assessment or can be requested at written exams using digital slides when students’ skills are tested.
Dedicated software help random test orders for students sitting side-by-side at exams and allows automated evaluation of multi-choice test results. In addition, computer technology extends teaching into a multi-partner online consultation. Any student can ask for control over the teaching software and specify a question to all, including the teacher, as opposed to one-by-one hidden communication with the teacher over a traditional light microscope.
In September 2007 the 1st Department of Pathology and Experimental Cancer Research at Semmelweis University, became the first (as far as we are aware) to start teaching histopathology for medical students by exclusively using a digital slide system in a 40-computer facility. Since then, students can access the 200 slides of the histopathology curriculum freely through the www.pathonet.com website, generating over 100,000 page loads per year by ~350 students. A survey showed that 97% of students regularly access this slide repository, particularly before exam periods.
Digital slide collections of national and international slide seminars for self-assessment training and external quality assurance (EQA) are also accessible through this site.
The EQA scheme run by QualiCont, a non-profit-making EQA organisation in Hungary, also utilises digital slide/smear based systems to test interpretational skills in haematopathology and cervix cytology regularly.
The College of American Pathologists (www.cap.org) also offers online digital slide programs in proficiency testing schemes for general surgical pathology and subspecialties such as dermatopathology or fine needle aspiration cytology.
A web-based digital atlas of breast histopathology (www.webmicroscope.net/breastatlas) has also proved to be an excellent tool for both graduate and postgraduate education. Digital microscopy can also assist in virtual tumour banking, such as in TuBaFrost (The European human frozen tumour tissue bank) by offering a digital slide catalogue of tumour samples stored at distant laboratories.
Accumulating knowledge on diseases has made pathology a highly sub-specialised discipline. In a country of ~10 million people such as Hungary, there are only few centres cover rare subspecialties, such as soft tissue-, haemato- or neuropathology.
Additionally, many laboratories are understaffed and/or under-equipped and all need regular external support through consultation. However, the sharing of glass slides for consultation, training sessions or slide seminars, creates substantial logistic problems. Sending glass slides via postal services carries the inherent risk of losing or damaging them and jeopardising patients' chances of getting the right diagnosis and treatment in time.
Dynamic telepathology using digital slides and dedicated software tools can solve these problems by supporting prompt consultations with experts without spatial or temporal restrictions. Digital consultation is now supported not only by wired computer systems but also by mobile tools like tablets and smartphones.
Thus, telepathology is a way of 'cloning expert pathologists', by allowing them to contribute to several laboratories from any location. Therefore, remote pathology can offer equal opportunities both for patients in rural areas though easy access to specialist consultations by pathologist, and for professionals of restricted mobility, including the physically disabled and those caring for children at home.
Automated image analysis, brightfield and FISH
In biomarker detection, signal intensity is proportional with the concentration of the targeted molecule; therefore, digital slides can be used for automated biomarker quantification to reduce the subjective bias of empirical scoring. However, due to the biochemical complexity of cells and difficulties of standardising tissue processing, calibrated absolute controls for tissue molecules are not yet available. Therefore, concentrations can only be assessed semi-quantitatively, which is still appropriate for performing comparative studies, e.g. correlating biomarker levels with clinical behaviour (tumour grade, survival etc.).
Initial colour separation based on deconvolution helps discriminate, for example, nuclear haematoxylin counterstaining from the chromogenic signals of immunohistochemistry. Several integrated scanning and image analysis algorithms are available for automated examination of brightfield and fluorescing signals within the QuantCenter package.
PatternQuant software permits automated identification and demarcation of morphological and functional units within tissues, such as glands, stromal areas, or hyperplastic or abnormally arranged epithelial tumour nests after preliminary training on shape, size and texture of targeted structures (Figure 4).
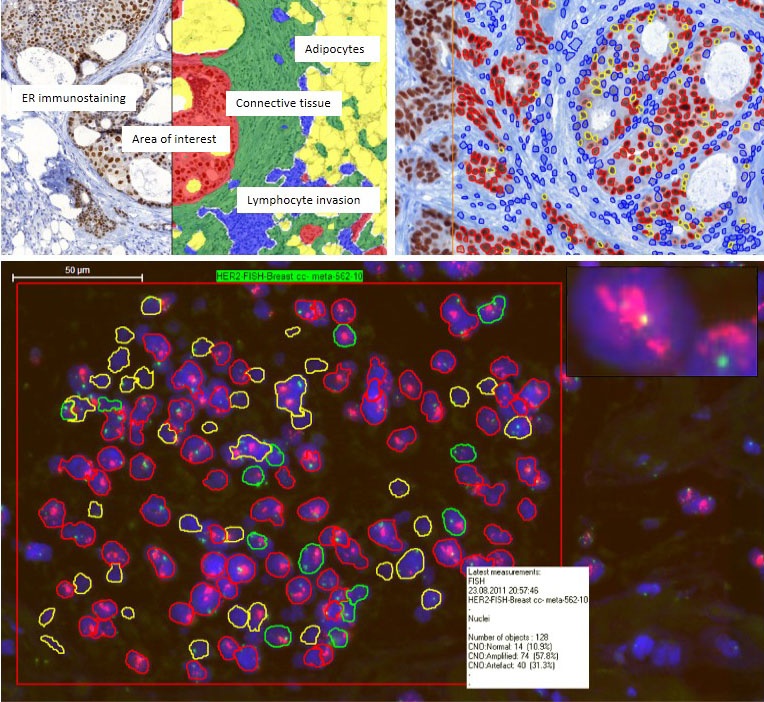
Then, selected regions can be further analysed using software tools dedicated for more specific tasks. Histoquant, a general image analysis program, is useful to determine area proportions of cytoplasmic immuno-staining reactions.
The CellQuant algorithm is most appropriate for counting positive cell numbers based on automated separation of individual cells, either accumulated in groups (e.g. tumour nests) or dispersed, such as inflammatory cells infiltrating tumours and their microenvironment.
The NuclearQuant program allows automated image analysis of nuclear signals, such as hormone receptors (ER/Pr), cell cycle regulation molecules (Ki67, cyclins etc.) or transcription factors (e.g. TTF1). Cell membrane associated signals, such as HER2, can be tested using the MembraneQuant software, which can automatically subclassify tumour cells into negative, 1+, 2+ and 3+ categories and determine their proportions according to the intensity and continuity of membrane positivity.
The DensitoQuant program is more focused on signal quantification by resulting in a H(histo)-Score, based on preliminary setting up the thresholds of four intensity ranges and multiplying intensity values with the frequency of positive cells within the target area.
Genomic in situ hybridisation results can be analysed by using the CISHQuant program if a chromogenic method was used or with the FISHQuant program for fluorescence-based detection of specific DNA regions. All genomic signals are particulate, thus they can be quantified with high precision using multichannel, multilayer scanning digital microscopy.
The algorithms classify cells based on the proportion and spatial localisation of nuclear gene and chromosome signals depending on the type of genetic test. These include numerical aberrations such as polysemy, aneusomy, amplification or loss, and structural aberrations such as translocations, or fusions with different algorithms used for fusion or break-apart probes. Results generated by the Quant programs are exported back to the XLS patient database and can serve for statistical and post-processing analysis including the generation of scatter plots, graphs and galleries.
Tissue microarrays (TMA), carrying tens or hundreds of small tissue cores in paraffin blocks, allow high throughput research of biomarkers, e.g. of cancer development and progression. The large amount of alphanumerical and image data can be handled efficiently and safely by using digital slides and database supported workflow and analysis within the TMA Module software package.
Utilisation of automated image analysis in such TMA projects carries high potential in cancer research. Since image data and the integrated measurements and annotations are permanent (including FISH), validation of datasets by other pathologists for quality assurance purposes is straightforward and easy.
The future of digital pathology
By taking advantage of the multi-functionality of computer technology digital microscopy allows rapid access to and analysis of information embedded in stained slides that can boost the efficiency of a pathology service. Digital pathology offers strong support in the challenge of coping with the growing workload, sub-specialisation and increasing expectations that clinicians and patients impose on a roughly constant number of pathology staff.
Digital slide-based histopathology allows system integration, which serve standardisation efforts through improving sample traceability and the reproducibility and accuracy of diagnostic reporting. These attributes are essential for quality management of the pathology workflow and support quality assurance initiatives. System integration involves network linking of procedures and sample data of all units within histopathology laboratories for tracing of diagnostic samples/slides from entry to reporting and monitoring indicators of quality.
The feasibility of using digital microscopy for diagnostic purposes is a highly complex issue determined by technical, professional and medico-legal aspects. These are related to scanning/server and network speed, image quality, storage space, appropriate backup system, the quality of workstation and graphic interface, technically trained professionals, internal method validation, ease of communication with the hospital database and secure data transfer protecting patients’ rights.
The general acceptance of digital pathology is supported by recent development in high-speed and capacity scanners, servers, networks and hospital database information (HIS) systems, and efforts to standardise digital slide file formats (DICOM).
Therefore, digital pathology has a great potential to be the driving force of modernising histopathology to improve patient care in the near future.
Source: Béla Molnár MD DSc
03.09.2017