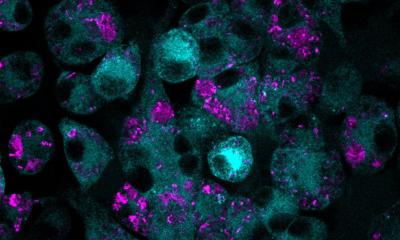
© Christian Urban, Kiel University
News • Rapid emergence of antibiotic resistance
How P. aeruginosa becomes resistant during CF treatment
Antibiotic-resistant pathogens pose one of the greatest threats to public health worldwide. In the near future, harmless bacterial infections may no longer be treatable and may again become the most common non-natural cause of death.
At the same time, the available repertoire of antibacterial agents is becoming increasingly smaller as resistance rates rise. The basic mechanisms of resistance evolution, i.e. the adaptation of a disease germ to the mode of action of a drug, have been well researched experimentally. How such loss of drug susceptibility develops in individual patients in the course of standard antibiotic therapy has not yet been studied. A research team from the department of Evolutionary Ecology and Genetics at Kiel University has now investigated the evolution of resistance in the pathogen Pseudomonas aeruginosa during antibiotic treatment in a cohort of cystic fibrosis patients as an example.
For the first time, they researched to what extent resistance evolves during a single course of antibiotic therapy. The researchers now found that resistance rapidly emerged in about one third of the patients. These findings were obtained within the Leibniz ScienceCampus "EvoLUNG" and the Cluster of Excellence "Precision Medicine in Chronic Inflammation" (PMI) and were recently published in the Journal of Antimicrobial Chemotherapy.
Cystic fibrosis lung infection as an example
In every third patient the pathogen adapted surprisingly quickly to the administered drugs and antibiotic resistance evolved within one to three days
Leif Tüffers
In the study, the research team focused on a small cohort of cystic fibrosis patients. The disease, which is uncurable to date, is caused by impaired cellular water transport and leads to viscous body secretions and associated dysfunction of numerous organs. It affects the respiratory tract and lungs in particular. Frequent infections in adult patients are mainly caused by Pseudomonas. As a result, they have to be treated with antibiotics frequently or even permanently.
In order to assess whether resistance evolves in the course of a single standard treatment with two or more antibiotics, the scientists examined the bronchial secretions of the patients daily and isolated Pseudomonas bacteria. This enabled them to investigate how the bacteria adapted to therapy over a period of 14 days. "In every third patient the pathogen adapted surprisingly quickly to the administered drugs and antibiotic resistance evolved within one to three days," summarised Dr Leif Tüffers, a researcher in the Department of Evolutionary Ecology and Genetics. "The rapid evolution of resistance mainly affected newly administered antibiotics from the betalactam drug class," continued Tüffers.
Findings match evolutionary experiments

© Christian Urban, Kiel University
These findings were obtained in real time on patients during clinical routine therapy for the first time. They generally correspond well with experimental observations from previous laboratory experiments with the Pseudomonas pathogen. Although resistances evolve faster in the laboratory, sometimes within a few hours, the bacteria in the patient's body grow much more slowly. To date, it is not entirely clear how exactly bacteria in the patients achieve such rapid evolution of resistance to the betalactam antibiotics, including penicillin. "It is possible that this rapid adaptation to the drug occurs as a result of spontaneous, new mutations in resistance genes," said Tüffers.
In current research within the Cluster of Excellence PMI, the scientists now investigate the clinical applicability of evolution-based treatment strategies, which proved successful under laboratory conditions in the past. "The processes of resistance evolution are often comparable, regardless of whether it is a laboratory experiment with a specific single bacterium or the treatment of a bacterial infection in patients," said Professor Hinrich Schulenburg, head of the Department of Evolutionary Ecology and Genetics at Kiel University. "We therefore want to investigate in future studies to what extent, for example, the principle of collateral sensitivity, in which the formation of resistance to one antibiotic increases sensitivity to a second drug, can be used to treat lung infections," continued Schulenburg.
Source: Kiel University
25.11.2019