Epidemics
Open Science to track virus outbreaks
In recent years, we have witnessed multiple epidemics of viral diseases such as Ebola or Zika. Rapid targeted intervention is key to containment. Real-time data integration and analysis can help public health authorities to maximize efficacy of intervention strategies. Dr. Richard Neher from the Max Planck Institute for Developmental Biology, Germany and Dr. Trevor Bedford from the Fred Hutchinson Cancer Research Center, Seattle, USA, won Phase 1 of the Open Science Prize Challenge for their proposal to develop an online platform for real-time evolutionary and epidemiological analysis. The results will be visualized using interactive web applications that make actionable results accessible.
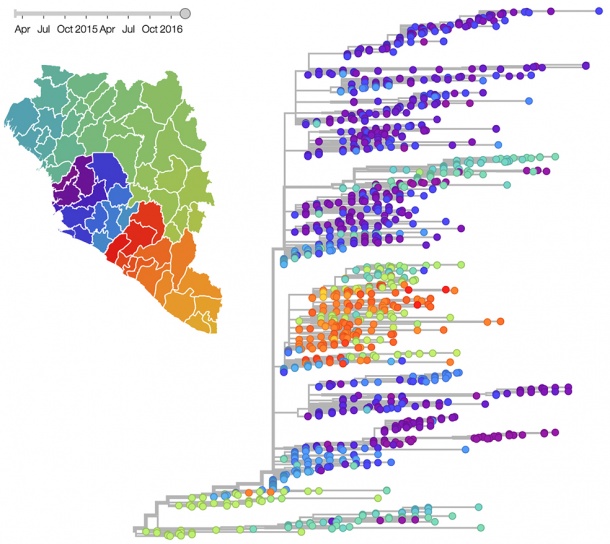

Viral pathogens are a global threat to public health. Influenza causes an estimated 250,000 to 500,000 deaths per year, and recent outbreak of Zika virus believed to be on track to cause tens of thousands of birth defects. Public health officials can only develop effective intervention strategies if the epidemiology and evolution of the viral infections is well understood. Genome sequencing provides excellent insight into the evolutionary and epidemiological dynamics of such viruses. To allow better, more targeted interventions, sequencing data from different groups, scientists or hospitals need to be analyzed in a timely fashion to maximize their value for public health.
This can only be achieved by rapid data sharing. Dr. Neher and Dr. Bedford have thus developed a bioinformatics tool that reconstructs a genealogical tree of the viruses by the mutations they randomly accumulate. “The tool pools data between sampling locations and research groups. A new sequence can be integrated into our output within 24 hours”, explains Neher. The software is already implemented for influenza (www.nextflu.org) and will now be extended to other viruses.
The platform will be publicly available on www.nextstrain.org and the source code released under a General Public License in order to provide software containers that can also be used by other scientists and rapidly adapted to different diseases.
The tool will infer geographical regions that are sources in driving virus circulation, which can help public health officials to target containment effort such as mosquito control. In addition, the tool will enable tracing of transmission chains, which is vital for disease eradication. Additionally, mutations in the virus genome will be analyzed to identify mutations that are suspected to change the virus transmissibility or pathogenicity.
The platform also aims at motivating research groups to share their data rapidly. “I think that outbreak response is an area in which open science and data sharing can make a huge difference. We aim to have a platform that promotes insights from data without treading on the original data producer’s publication priority”, says Bedford.
The Open Science Prize is a partnership between the Wellcome Trust, the US National Institutes of Health (NIH) and the Howard Hughes Medical Institute to unleash the power of open content and data to advance biomedical research and its application for health benefit. This first round of the Prize consists of a two-phase competition. For the first phase, international teams competed for funding to take new ideas for products or services to the prototype stage, or to further develop an existing early-stage prototype. Prizes of $80,000 each were awarded today to successful teams. In the second phase, the phase I prize recipient judged to have the prototype with the greatest potential to advance open science will receive a prize of $230,000.
Source: Max Planck Institute for Developmental Biology
13.05.2016