Image credit: Andrew H. Song, Brigham and Women's Hospital
News • Three-dimensional tissue processing
Pathology performs leap into 3D with AI
Human tissue is intricate, complex and, of course, three-dimensional. But the thin slices of tissue that pathologists most often use to diagnose disease are two-dimensional, offering only a limited glimpse at the tissue’s true complexity.
There is a growing push in the field of pathology toward examining tissue in its three-dimensional form. But 3D pathology datasets can contain hundreds of times more data than their 2D counterparts, making manual examination infeasible.
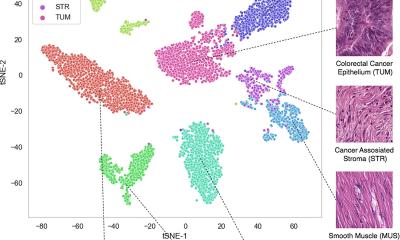
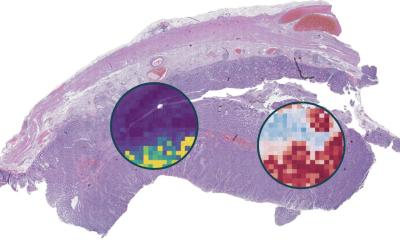
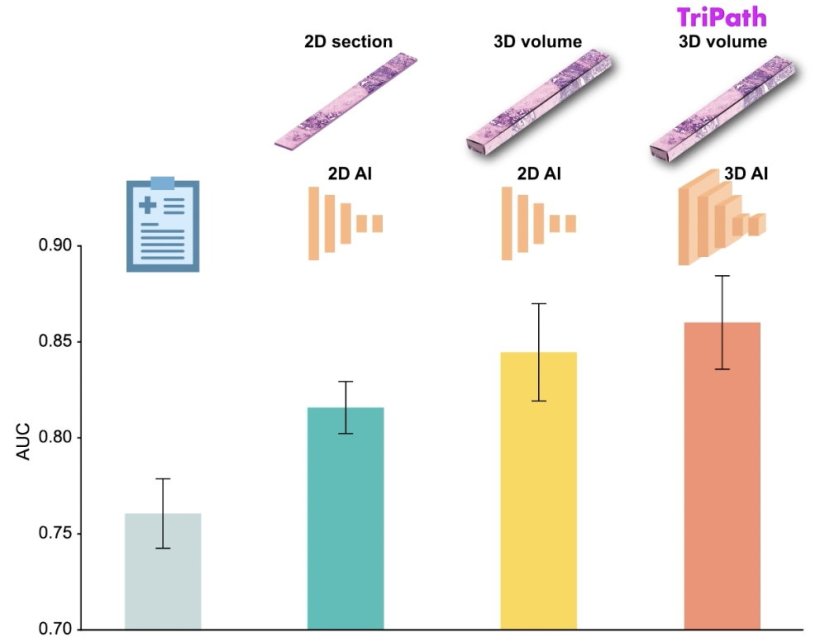
In a new study, researchers from Mass General Brigham and their collaborators present Tripath: new, deep learning models that can use 3D pathology datasets to make clinical outcome predictions. In collaboration with the University of Washington, the research team imaged curated prostate cancer specimens, using two 3D high-resolution imaging techniques. The models were then trained to predict prostate cancer recurrence risk on volumetric human tissue biopsies. By comprehensively capturing 3D morphologies from the entire tissue volume, Tripath performed better than pathologists and outperformed deep learning models that rely on 2D morphology and thin tissue slices. Results are published in Cell.
Our approach underscores the importance of comprehensively analyzing the whole volume of a tissue sample for accurate patient risk prediction
Andrew H. Song
While the new approach needs to be validated in larger datasets before it can be further developed for clinical use, the researchers are optimistic about its potential to help inform clinical decision making. “Our approach underscores the importance of comprehensively analyzing the whole volume of a tissue sample for accurate patient risk prediction, which is the hallmark of the models we developed and only possible with the 3D pathology paradigm,” said lead author Andrew H. Song, PhD, of the Division of Computational Pathology in the Department of Pathology at Mass General Brigham.
Recommended article

News • Research on large foundation models
Reducing bias in pathology AI
A study highlights performance differences in computational pathology systems, depending on demographic profiles associated with histology images. The researchers also propose a way to fix this bias.
“Using advancements in AI and 3D spatial biology techniques, Tripath provides a framework for clinical decision support and may help reveal novel biomarkers for prognosis and therapeutic response,” said co-corresponding author Faisal Mahmood, PhD, of the Division of Computational Pathology in the Department of Pathology at Mass General Brigham.
“In our prior work in computational 3D pathology, we looked at specific structures such as the prostate gland network, but Tripath is our first attempt to use deep learning to extract sub-visual 3D features for risk stratification, which shows promising potential for guiding critical treatment decisions,” said co-corresponding author Jonathan Liu, PhD, at the University of Washington.
Source: Mass General Brigham
10.05.2024